DNA MODIFICATION PATHWAY (PW:0001340)
Description
Epigenetic changes, the modification and remodeling of chromatin, play a key role in the differential expression of genes. Chromatin modification and remodeling directly affect the relative relaxation or compaction of chromatin and thus, the extent to which DNA replication, transcription, damage response and repair, associated RNA processing and splicing are promoted or are silenced, respectively. The basic unit of chromatin is the nucleosome - it consists of 145-147 base pairs of DNA wrapped around an octameric structure formed by core histones. Repeating nucleosomes assemble into higher-order structures stabilized by a linker histone. Chromatin modification involves the methylation of cytosine residues in DNA and the many ways in which the histone residues are modified. Histone modifications mostly targets the histone tails which extend from the core fold of the nucleosome and thus, are accessible for modifications and interactions. The modification pathways bring together the enzymes that catalyze the modification and those that catalyze its removal or change, known as 'writer' and 'erasers', respectively, the proteins whose particular domains recognize the modification, known as the 'readers' and the various other interactors or regulators. The remodeling of chromatin involves the ATP-dependent eviction, sliding, deposition or exchange of entire nucleosomes. It is carried out by four protein complexes representing four remodeling pathways with distinct functions. The replication-dependent and independent assembly of nucleosomes is mediated by histone chaperone proteins. Epigenetic changes are heritable and independent of DNA sequence. However, DNA sequence variation can affect the function of players involved and thus, of epigenetic outcomes. An intimate dialog exists between the pathways of chromatin modification and remodeling to finely tune the epigenetic responses. The interaction of chromatin modifying enzymes and readers with non-coding RNA, particularly long non-coding RNA (lncRNA), further modulates the epigenetic landscape and the outcomes of regulated gene expression. Chromatin modifying enzymes and readers are target of drug development, primarily for cancer treatment. Several members are also found mutated in a number of human cancers. The DNA modification pathway is presented here.
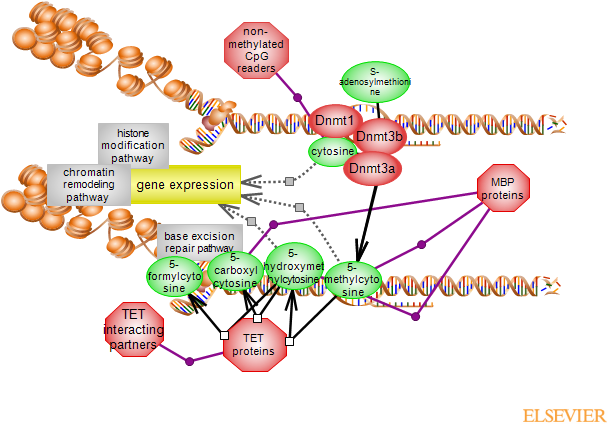
DNA methylation occurs within CpG genomic dinucletodies and is found on up to 80% of the genome. However, non-methylated regions, known as CpG islands (CGI) are found in ~70-80% of vertebrate promoters. DNA methylation controls the X-chromosome inactivation and the expression of imprinted genes which are important regulators of development, among others. It generally represses gene expression and serves as an important marker that influences not only chromatin changes, remodeling and transcription but also splicing. Three DNA methyltransferase (DNMTs) carry out the reaction; DNMT1 is considered a maintenance methyltransferase and the other two mostly function as de novo methyltransferases. DNMT1 is responsible for reinstating the methylation pattern to daughter strand during replication. S-adenosylmethionine (SAM) intermediate generated in the methionine cycle metabolic pathway is the methyl donor and the product of the reaction is 5-methylcytosine (5mC). The methylated cytosines are recognized by methyl binding proteins (MBPs) or factors which can then recruit other chromatin modifiers, modelers and regulators. The methyl binding proteins can be classified into three structural families: methyl-CpG binding domain (MBD), the zinc finger family and the SET and RING finger-associated (SRA) family. Of these, MBD1, 2 and 4 and MeCP2 are members of MBD family, Zbtb33 known as Kaiso, Zbtb4, Zbtb38 and Zfp57 are members of the zinc family and Uhrf1 and 2 are members of SRA family. The are no known demethylase enzymes that directly act upon 5mC. 5mC can be converted to 5-hydroxymethylcytosine (5hmC) which can be further oxidized to 5-formylcytosine (5fC) and 5-carboxycytosine (5caC) by the members of ten-eleven translocation (TET) family. The proteins are 2-oxoglutarate and Fe(II)-dependent dioxygenases. Of the three TET proteins, Tet2 and 3 are ubiquitously expressed; the expression of Tet 1 is confined to embryonic cells. Some of the 5mC intermediates interact with members of the MBPs protein families and they can be more than just posts along the demethylation route. 5hmC for instance, has been shown to have both transcriptional activation and silencing roles. There are no known enzymes catalyzing the removal of formyl or carbonyl groups from 5fC or5caC, but they may be substrates for glycosylases and subsequent targeting by the base excision repair (BER) pathway. Tet enzymes are partners for proteins with roles in shaping the state of chromatin and the regulation of gene expression, modulating but also extending the function of Tet proteins beyond 5mC modification. There is a complex interplay between DNA methylation and histone modification, particularly histone lysine methylation. Proteins that bind methylated CpG interact with histones and histone modifying enzymes. In addition, proteins with the ZF-CxxC (cysteine-rich zinc finger-CxxC) domain, bind non-methylated CpG to modulate histone methylation. ZF-CxxC members group together histone and DNA modifying enzymes, components of modifying complexes such as COMPASS. The DNA methylation pathway is frequently altered in several conditions, including cancer. Hypermethylation of tumor suppressor and hypomethylation of oncogene gene promoters have been reported.To link to the Ontology Report for annotations, GViewer and download, click here ...(less)
Pathway Diagram:
Genes in Pathway:
G
B
Cxxc1
CXXC finger protein 1
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr15:57,715,783...57,733,286
G
N
Cxxc1
CXXC finger protein 1
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004624778:13,908,430...13,915,273
G
G
CXXC1
CXXC finger protein 1
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr18:30,668,879...30,674,841
G
P
CXXC1
CXXC finger protein 1
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr 1:99,934,493...99,940,125
G
S
Cxxc1
CXXC finger protein 1
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004936497:12,842,569...12,847,952
G
D
CXXC1
CXXC finger protein 1
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr 7:78,591,288...78,596,642
G
B
CXXC1
CXXC finger protein 1
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr18:43,662,843...43,668,825
G
C
Cxxc1
CXXC finger protein 1
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004955402:35,335,520...35,340,891
G
R
Cxxc1
CXXC finger protein 1
ISO
RGD
PMID:23697932 RGD:9479164
NCBI chr18:70,088,407...70,093,786
G
M
Cxxc1
CXXC finger protein 1
ISO
RGD
PMID:23697932 RGD:9479164
NCBI chr18:74,349,283...74,354,564
G
H
CXXC1
CXXC finger protein 1
TAS
RGD
PMID:23697932 RGD:9479164
NCBI chr18:50,282,347...50,287,692
G
B
Dnmt1
DNA methyltransferase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 8:17,168,919...17,213,593
G
N
Dnmt1
DNA methyltransferase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004624828:2,316,042...2,358,866
G
G
DNMT1
DNA methyltransferase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 6:9,199,006...9,263,890
G
P
DNMT1
DNA methyltransferase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 2:68,981,564...69,040,364
G
S
Dnmt1
DNA methyltransferase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004936659:520,181...565,496
G
D
DNMT1
DNA methyltransferase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr20:50,872,213...50,928,352
G
B
DNMT1
DNA methyltransferase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr19:9,681,847...9,745,334
G
C
Dnmt1
DNA methyltransferase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004955495:1,465,529...1,505,379
G
R
Dnmt1
DNA methyltransferase 1
ISO
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr 8:27,716,797...27,763,405
G
M
Dnmt1
DNA methyltransferase 1
ISO
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr 9:20,818,501...20,871,084
G
H
DNMT1
DNA methyltransferase 1
TAS
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr19:10,133,346...10,194,953
G
B
Dnmt3a
DNA methyltransferase 3 alpha
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 7:24,482,595...24,583,861
G
N
Dnmt3a
DNA methyltransferase 3 alpha
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004624738:7,481,084...7,581,332
G
G
DNMT3A
DNA methyltransferase 3 alpha
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr14:82,313,414...82,427,170
G
P
DNMT3A
DNA methyltransferase 3 alpha
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 3:113,501,972...113,609,148
G
S
Dnmt3a
DNA methyltransferase 3 alpha
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004936493:6,963,406...7,013,256
G
D
DNMT3A
DNA methyltransferase 3 alpha
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr17:19,489,524...19,563,074
G
B
DNMT3A
DNA methyltransferase 3 alpha
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr2A:25,229,593...25,344,733
G
C
Dnmt3a
DNA methyltransferase 3 alpha
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004955469:7,264,431...7,367,059
G
R
Dnmt3a
DNA methyltransferase 3 alpha
ISO
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr 6:32,507,316...32,621,678
G
M
Dnmt3a
DNA methyltransferase 3A
ISO
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr12:3,851,559...3,964,442
G
H
DNMT3A
DNA methyltransferase 3 alpha
TAS
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr 2:25,227,874...25,342,590
G
N
Dnmt3b
DNA methyltransferase 3 beta
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004624842:6,450,972...6,505,929
G
G
DNMT3B
DNA methyltransferase 3 beta
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 2:37,128,635...37,176,033
G
P
DNMT3B
DNA methyltransferase 3 beta
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr17:36,345,715...36,386,076
G
S
Dnmt3b
DNA methyltransferase 3 beta
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004936485:19,547,836...19,570,716
G
D
DNMT3B
DNA methyltransferase 3 beta
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr24:22,096,386...22,123,378
G
B
DNMT3B
DNA methyltransferase 3 beta
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr20:29,049,993...29,096,957
G
C
Dnmt3b
DNA methyltransferase 3 beta
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004955422:28,380,595...28,427,012
G
R
Dnmt3b
DNA methyltransferase 3 beta
ISO
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr 3:162,590,777...162,629,313
G
M
Dnmt3b
DNA methyltransferase 3B
ISO
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr 2:153,491,332...153,529,650
G
H
DNMT3B
DNA methyltransferase 3 beta
TAS
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr20:32,762,385...32,809,356
G
B
Ebf1
EBF transcription factor 1
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 9:27,435,767...27,822,514
G
N
Ebf1
EBF transcription factor 1
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004624733:30,327,541...30,735,432
G
G
EBF1
EBF transcription factor 1
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr23:61,097,268...61,502,784
G
P
EBF1
EBF transcription factor 1
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr16:64,357,999...64,753,188
G
S
Ebf1
EBF transcription factor 1
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004936515:4,266,419...4,643,702
G
D
EBF1
EBF transcription factor 1
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 4:51,378,314...51,765,768
G
B
EBF1
EBF transcription factor 1
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 5:154,091,337...154,492,474
G
C
Ebf1
EBF transcription factor 1
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004955408:12,735,893...13,121,700
G
R
Ebf1
EBF transcription factor 1
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr10:29,597,221...29,990,571
G
M
Ebf1
early B cell factor 1
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr11:44,502,738...44,898,923
G
H
EBF1
EBF transcription factor 1
TAS
RGD
PMID:24825349 RGD:8695944
NCBI chr 5:158,695,920...159,099,916
G
B
Kdm2a
lysine demethylase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr 2:3,259,066...3,328,896
G
N
Kdm2a
lysine demethylase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004624767:19,247,009...19,365,520
G
G
KDM2A
lysine demethylase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr 1:7,004,109...7,152,072
G
P
KDM2A
lysine demethylase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr 2:5,238,690...5,357,799
G
S
Kdm2a
lysine demethylase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004936599:2,466,427...2,576,623
G
D
KDM2A
lysine demethylase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr18:50,178,511...50,294,097
G
B
KDM2A
lysine demethylase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr11:62,470,549...62,616,135
G
C
Kdm2a
lysine demethylase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004955422:18,201,243...18,308,163
G
R
Kdm2a
lysine demethylase 2A
ISO
RGD
PMID:23697932 RGD:9479164
NCBI chr 1:211,041,859...211,125,749
G
M
Kdm2a
lysine (K)-specific demethylase 2A
ISO
RGD
PMID:23697932 RGD:9479164
NCBI chr19:4,366,172...4,448,749
G
H
KDM2A
lysine demethylase 2A
TAS
RGD
PMID:23697932 RGD:9479164
NCBI chr11:67,119,263...67,258,082
G
B
Kdm2b
lysine demethylase 2B
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr16:30,834,088...30,960,270
G
N
Kdm2b
lysine demethylase 2B
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004624747:21,922,249...22,047,246
G
G
KDM2B
lysine demethylase 2B
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr11:116,805,541...116,957,236
G
P
KDM2B
lysine demethylase 2B
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr14:31,057,121...31,203,093
G
S
Kdm2b
lysine demethylase 2B
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004936558:2,771,420...2,897,694
G
D
KDM2B
lysine demethylase 2B
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr26:7,625,857...7,746,049
G
B
KDM2B
lysine demethylase 2B
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr12:119,027,633...119,183,779
G
C
Kdm2b
lysine demethylase 2B
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004955482:6,774,021...6,885,113
G
R
Kdm2b
lysine demethylase 2B
ISO
RGD
PMID:23697932 RGD:9479164
NCBI chr12:39,224,422...39,362,126
G
M
Kdm2b
lysine (K)-specific demethylase 2B
ISO
RGD
PMID:23697932 RGD:9479164
NCBI chr 5:123,008,727...123,127,333
G
H
KDM2B
lysine demethylase 2B
TAS
RGD
PMID:23697932 RGD:9479164
NCBI chr12:121,408,461...121,582,279
G
B
Kmt2a
lysine methyltransferase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr 8:39,336,343...39,416,125
G
N
Kmt2a
lysine methyltransferase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004624784:13,824,507...13,912,672
G
G
KMT2A
lysine methyltransferase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr 1:109,819,748...109,912,763
G
P
KMT2A
lysine methyltransferase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr 9:45,743,566...45,828,559
G
S
Kmt2a
lysine methyltransferase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004936542:3,480,071...3,566,823
G
D
KMT2A
lysine methyltransferase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr 5:15,212,559...15,302,825
G
B
KMT2A
lysine methyltransferase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr11:113,273,198...113,364,404
G
C
Kmt2a
lysine methyltransferase 2A
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004955412:19,586,435...19,670,149
G
R
Kmt2a
lysine methyltransferase 2A
ISO
RGD
PMID:23697932 RGD:9479164
NCBI chr 8:54,013,547...54,089,219
G
M
Kmt2a
lysine (K)-specific methyltransferase 2A
ISO
RGD
PMID:23697932 RGD:9479164
NCBI chr 9:44,714,652...44,793,492
G
H
KMT2A
lysine methyltransferase 2A
TAS
RGD
PMID:23697932 RGD:9479164
NCBI chr11:118,436,492...118,526,832
G
N
Kmt2c
lysine methyltransferase 2C
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004624800:4,510,669...4,785,909
G
G
KMT2C
lysine methyltransferase 2C
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr21:120,299,295...120,587,843
G
P
KMT2C
lysine methyltransferase 2C
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr18:4,993,213...5,279,668
G
S
Kmt2c
lysine methyltransferase 2C
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004936527:7,310,885...7,508,032
G
D
KMT2C
lysine methyltransferase 2C
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr16:15,962,045...16,117,601
G
B
KMT2C
lysine methyltransferase 2C
ISO
RGD PMID:23697932 RGD:9479164
NCBI chr 7:143,714,716...144,016,188
G
C
Kmt2c
lysine methyltransferase 2C
ISO
RGD PMID:23697932 RGD:9479164
NCBI chrNW_004955491:6,245,408...6,420,535
G
R
Kmt2c
lysine methyltransferase 2C
ISO
RGD
PMID:23697932 RGD:9479164
NCBI chr 4:10,353,698...10,755,965
G
M
Kmt2c
lysine (K)-specific methyltransferase 2C
ISO
RGD
PMID:23697932 RGD:9479164
NCBI chr 5:25,476,793...25,703,853
G
H
KMT2C
lysine methyltransferase 2C
TAS
RGD
PMID:23697932 RGD:9479164
NCBI chr 7:152,134,925...152,436,003
G
B
LOC100977775
zinc finger protein 707
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 6:29,476,459...29,485,480
G
G
LOC103222426
methyl-CpG-binding domain protein 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr18:30,675,419...30,690,111
G
B
Mbd1
methyl-CpG binding domain protein 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr15:57,666,542...57,681,023
G
N
Mbd1
methyl-CpG binding domain protein 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004624778:13,892,179...13,907,576
G
P
MBD1
methyl-CpG binding domain protein 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 1:99,848,193...99,917,347
G
S
Mbd1
methyl-CpG binding domain protein 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004936497:12,848,918...12,868,504
G
D
MBD1
methyl-CpG binding domain protein 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 7:78,597,688...78,611,303
G
B
MBD1
methyl-CpG binding domain protein 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr18:43,647,395...43,662,266
G
C
Mbd1
methyl-CpG binding domain protein 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004955402:35,320,755...35,334,751
G
R
Mbd1
methyl-CpG binding domain protein 1
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr18:70,145,093...70,159,744
G
M
Mbd1
methyl-CpG binding domain protein 1
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr18:74,400,676...74,415,808
G
H
MBD1
methyl-CpG binding domain protein 1
TAS
RGD
PMID:23324617 RGD:8695954
NCBI chr18:50,266,885...50,281,767
G
N
Mbd2
methyl-CpG binding domain protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004624778:17,880,480...17,959,879
G
G
MBD2
methyl-CpG binding domain protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr18:26,714,610...26,785,028
G
P
MBD2
methyl-CpG binding domain protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 1:103,683,077...103,768,372
G
S
Mbd2
methyl-CpG binding domain protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004936497:9,512,350...9,619,250
G
D
MBD2
methyl-CpG binding domain protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 1:21,232,260...21,283,687
G
B
MBD2
methyl-CpG binding domain protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr18:47,534,356...47,609,481
G
C
Mbd2
methyl-CpG binding domain protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004955402:38,761,668...38,829,830
G
R
Mbd2
methyl-CpG binding domain protein 2
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr18:66,449,196...66,516,112
G
M
Mbd2
methyl-CpG binding domain protein 2
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr18:70,701,298...70,761,769
G
H
MBD2
methyl-CpG binding domain protein 2
TAS
RGD
PMID:23324617 RGD:8695954
NCBI chr18:54,151,606...54,224,669
G
B
Mbd4
methyl-CpG binding domain 4, DNA glycosylase
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 6:30,872,004...30,882,213
G
N
Mbd4
methyl-CpG binding domain 4, DNA glycosylase
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004624872:453,303...462,661
G
G
MBD4
methyl-CpG binding domain 4, DNA glycosylase
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr22:51,616,840...51,626,232
G
P
MBD4
methyl-CpG binding domain 4, DNA glycosylase
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr13:68,816,677...68,828,559
G
S
Mbd4
methyl-CpG binding domain 4, DNA glycosylase
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004936602:968,159...980,452
G
D
MBD4
methyl-CpG binding domain 4, DNA glycosylase
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr20:5,716,162...5,725,068
G
B
MBD4
methyl-CpG binding domain 4, DNA glycosylase
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 3:126,489,125...126,498,034
G
C
Mbd4
methyl-CpG binding domain 4, DNA glycosylase
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004955429:17,858,656...17,868,111
G
R
Mbd4
methyl-CpG binding domain 4 DNA glycosylase
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr 4:150,565,646...150,577,433
G
M
Mbd4
methyl-CpG binding domain protein 4
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr 6:115,817,658...115,830,361
G
H
MBD4
methyl-CpG binding domain 4, DNA glycosylase
TAS
RGD
PMID:23324617 RGD:8695954
NCBI chr 3:129,430,947...129,439,948
G
B
Mecp2
methyl-CpG binding protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr X:128,480,073...128,534,447
G
N
Mecp2
methyl-CpG binding protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004624946:620,581...685,339
G
G
MECP2
methyl-CpG binding protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr X:128,445,028...128,519,892
G
P
MECP2
methyl-CpG binding protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr X:124,735,523...124,789,063
G
S
Mecp2
methyl-CpG binding protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004936809:902,938...967,771
G
D
MECP2
methyl-CpG binding protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr X:121,866,721...121,876,088
G
B
MECP2
methyl-CpG binding protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr X:143,571,864...143,647,192
G
C
Mecp2
methyl-CpG binding protein 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004955580:674,014...737,586
G
R
Mecp2
methyl CpG binding protein 2
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr X:156,932,481...156,995,981
G
M
Mecp2
methyl CpG binding protein 2
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr X:73,070,198...73,129,296
G
H
MECP2
methyl-CpG binding protein 2
TAS
RGD
PMID:23324617 RGD:8695954
NCBI chr X:154,021,573...154,097,717
G
B
Nanog
Nanog homeobox
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 6:24,378,513...24,386,158
G
N
Nanog
Nanog homeobox
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004624860:5,026,203...5,037,219
G
G
NANOG
Nanog homeobox
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr11:7,813,309...7,820,009
G
P
NANOG
Nanog homeobox
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 5:62,931,642...62,939,086
G
S
Nanog
Nanog homeobox
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004936892:181,834...187,691
G
D
NANOG
Nanog homeobox
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr27:37,262,558...37,266,449
G
B
NANOG
Nanog homeobox
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr12:7,913,037...7,919,715
G
C
Nanog
Nanog homeobox
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004955413:6,561,323...6,565,770
G
R
Nanog
Nanog homeobox
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr 4:157,615,687...157,623,061
G
M
Nanog
Nanog homeobox
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr 6:122,684,448...122,691,592
G
H
NANOG
Nanog homeobox
TAS
RGD
PMID:24825349 RGD:8695944
NCBI chr12:7,789,402...7,799,146
G
B
Ogt
O-linked N-acetylglucosamine (GlcNAc) transferase
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr X:58,824,800...58,867,294
G
N
Ogt
O-linked N-acetylglucosamine (GlcNAc) transferase
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004624903:1,863,110...1,912,299
G
G
OGT
O-linked N-acetylglucosamine (GlcNAc) transferase
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr X:61,351,047...61,393,541
G
P
OGT
O-linked N-acetylglucosamine (GlcNAc) transferase
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr X:57,521,091...57,558,467
G
S
Ogt
O-linked N-acetylglucosamine (GlcNAc) transferase
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004936762:99,335...137,833
G
D
OGT
O-linked N-acetylglucosamine (GlcNAc) transferase
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr X:55,821,227...55,857,597
G
B
OGT
O-linked N-acetylglucosamine (GlcNAc) transferase
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr X:60,798,219...60,840,948
G
C
Ogt
O-linked N-acetylglucosamine (GlcNAc) transferase
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004955475:11,062,258...11,112,252
G
R
Ogt
O-linked N-acetylglucosamine (GlcNAc) transferase
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr X:70,811,317...70,856,123
G
M
Ogt
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr X:100,683,617...100,727,957
G
H
OGT
O-linked N-acetylglucosamine (GlcNAc) transferase
TAS
RGD
PMID:24825349 RGD:8695944
NCBI chr X:71,533,104...71,575,892
G
B
Pparg
peroxisome proliferator activated receptor gamma
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 6:31,222,604...31,347,537
G
N
Pparg
peroxisome proliferator activated receptor gamma
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004624956:84,673...207,176
G
G
PPARG
peroxisome proliferator activated receptor gamma
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr22:48,262,743...48,407,415
G
P
PPARG
peroxisome proliferator activated receptor gamma
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr13:68,301,566...68,433,951
G
S
Pparg
peroxisome proliferator activated receptor gamma
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004936602:1,230,907...1,368,588
G
D
PPARG
peroxisome proliferator activated receptor gamma
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr20:6,134,618...6,265,481
G
B
PPARG
peroxisome proliferator activated receptor gamma
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 3:12,215,322...12,361,669
G
C
Pparg
peroxisome proliferator activated receptor gamma
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004955429:14,814,690...14,936,869
G
R
Pparg
peroxisome proliferator-activated receptor gamma
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr 4:150,095,743...150,221,104
G
M
Pparg
peroxisome proliferator activated receptor gamma
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr 6:115,337,828...115,467,365
G
H
PPARG
peroxisome proliferator activated receptor gamma
TAS
RGD
PMID:24825349 RGD:8695944
NCBI chr 3:12,287,368...12,434,344
G
B
Prdm14
PR/SET domain 14
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 1:147,762,841...147,772,982
G
N
Prdm14
PR/SET domain 14
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004624744:21,092,091...21,121,313
G
G
PRDM14
PR/SET domain 14
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 8:65,800,454...65,820,190
G
P
PRDM14
PR/SET domain 14
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 4:65,273,988...65,295,362
G
S
Prdm14
PR/SET domain 14
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004936496:14,789,443...14,820,967
G
D
PRDM14
PR/SET domain 14
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr29:19,203,798...19,220,149
G
B
PRDM14
PR/SET domain 14
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 8:66,578,674...66,599,360
G
C
Prdm14
PR/SET domain 14
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004955444:9,606,310...9,626,772
G
R
Prdm14
PR/SET domain 14
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 5:10,864,137...10,876,666
G
M
Prdm14
PR domain containing 14
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr 1:13,183,649...13,197,467
G
H
PRDM14
PR/SET domain 14
TAS
RGD
PMID:24825349 RGD:8695944
NCBI chr 8:70,051,651...70,071,252
G
B
Sin3a
SIN3 transcription regulator family member A
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 8:50,986,820...51,041,444
G
N
Sin3a
SIN3 transcription regulator family member A
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004624894:300,027...388,397
G
G
SIN3A
SIN3 transcription regulator family member A
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr26:7,971,208...8,060,888
G
P
SIN3A
SIN3 transcription regulator family member A
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 7:58,171,821...58,246,126
G
S
Sin3a
SIN3 transcription regulator family member A
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004936471:34,206,084...34,260,538
G
D
SIN3A
SIN3 transcription regulator family member A
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr30:38,231,844...38,306,665
G
B
SIN3A
SIN3 transcription regulator family member A
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr15:54,285,587...54,371,376
G
C
Sin3a
SIN3 transcription regulator family member A
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004955450:2,804,551...2,870,450
G
R
Sin3a
SIN3 transcription regulator family member A
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr 8:66,377,471...66,432,150
G
M
Sin3a
transcriptional regulator, SIN3A (yeast)
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr 9:56,979,271...57,035,652
G
H
SIN3A
SIN3 transcription regulator family member A
TAS
RGD
PMID:24825349 RGD:8695944
NCBI chr15:75,369,379...75,455,815
G
B
Spi1
Spi-1 proto-oncogene
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 5:65,264,663...65,283,117
G
N
Spi1
Spi-1 proto-oncogene
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004624767:1,376,649...1,404,382
G
G
SPI1
Spi-1 proto-oncogene
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 1:17,935,037...17,961,638
G
P
SPI1
Spi-1 proto-oncogene
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr 2:15,227,850...15,245,679
G
S
Spi1
Spi-1 proto-oncogene
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004936562:1,852,759...1,870,647
G
D
SPI1
Spi-1 proto-oncogene
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr18:42,253,849...42,269,497
G
B
SPI1
Spi-1 proto-oncogene
ISO
RGD PMID:24825349 RGD:8695944
NCBI chr11:47,301,245...47,326,891
G
C
Spi1
Spi-1 proto-oncogene
ISO
RGD PMID:24825349 RGD:8695944
NCBI chrNW_004955422:873,735...886,560
G
R
Spi1
Spi-1 proto-oncogene
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr 3:97,529,509...97,548,204
G
M
Spi1
Spi-1 proto-oncogene
ISO
RGD
PMID:24825349 RGD:8695944
NCBI chr 2:90,912,750...90,946,104
G
H
SPI1
Spi-1 proto-oncogene
TAS
RGD
PMID:24825349 RGD:8695944
NCBI chr11:47,354,860...47,378,547
G
B
Tet1
tet methylcytosine dioxygenase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr18:23,815,875...23,883,755
G
N
Tet1
tet methylcytosine dioxygenase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004624754:2,846,479...2,966,845
G
G
TET1
tet methylcytosine dioxygenase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 9:62,709,800...62,843,322
G
P
TET1
tet methylcytosine dioxygenase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr14:71,675,671...71,812,732
G
D
TET1
tet methylcytosine dioxygenase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 4:19,759,661...19,898,345
G
B
TET1
tet methylcytosine dioxygenase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr10:65,036,313...65,168,051
G
C
Tet1
tet methylcytosine dioxygenase 1
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004955425:20,767,406...20,857,165
G
R
Tet1
tet methylcytosine dioxygenase 1
ISO
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr20:25,766,806...25,839,598
G
M
Tet1
tet methylcytosine dioxygenase 1
ISO
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr10:62,640,349...62,723,242
G
H
TET1
tet methylcytosine dioxygenase 1
TAS
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr10:68,560,337...68,694,487
G
B
Tet2
tet methylcytosine dioxygenase 2
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 3:25,401,084...25,481,014
G
N
Tet2
tet methylcytosine dioxygenase 2
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004624830:3,446,331...3,543,450
G
G
TET2
tet methylcytosine dioxygenase 2
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 7:53,193,824...53,326,813
G
P
TET2
tet methylcytosine dioxygenase 2
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 8:116,413,253...116,546,976
G
S
Tet2
tet methylcytosine dioxygenase 2
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004936752:440,137...577,339
G
D
TET2
tet methylcytosine dioxygenase 2
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr32:26,021,745...26,145,109
G
B
TET2
tet methylcytosine dioxygenase 2
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chr 4:97,612,439...97,745,544
G
C
Tet2
tet methylcytosine dioxygenase 2
ISO
RGD PMID:22770212 PMID:24825349 RGD:8695943 RGD:8695944
NCBI chrNW_004955496:3,885,684...3,963,056
G
R
Tet2
tet methylcytosine dioxygenase 2
ISO
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr 2:224,662,654...224,746,819
G
M
Tet2
tet methylcytosine dioxygenase 2
ISO
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr 3:133,169,438...133,250,882
G
H
TET2
tet methylcytosine dioxygenase 2
TAS
RGD
PMID:22770212 PMID:24825349 RGD:8695943 , RGD:8695944
NCBI chr 4:105,145,875...105,279,803
G
B
Tet3
tet methylcytosine dioxygenase 3
ISO
RGD PMID:22770212 PMID:23697932 PMID:24825349 RGD:8695943 RGD:8695944 RGD:9479164
NCBI chr 6:62,766,910...62,859,977
G
N
Tet3
tet methylcytosine dioxygenase 3
ISO
RGD PMID:22770212 PMID:23697932 PMID:24825349 RGD:8695943 RGD:8695944 RGD:9479164
NCBI chrNW_004624749:29,371,674...29,494,785
G
G
TET3
tet methylcytosine dioxygenase 3
ISO
RGD PMID:22770212 PMID:23697932 PMID:24825349 RGD:8695943 RGD:8695944 RGD:9479164
NCBI chr14:33,170,046...33,293,128
G
P
TET3
tet methylcytosine dioxygenase 3
ISO
RGD PMID:22770212 PMID:23697932 PMID:24825349 RGD:8695943 RGD:8695944 RGD:9479164
NCBI chr 3:68,918,896...69,020,406
G
S
Tet3
tet methylcytosine dioxygenase 3
ISO
RGD PMID:22770212 PMID:23697932 PMID:24825349 RGD:8695943 RGD:8695944 RGD:9479164
NCBI chrNW_004936556:292,551...391,644
G
D
TET3
tet methylcytosine dioxygenase 3
ISO
RGD PMID:22770212 PMID:23697932 PMID:24825349 RGD:8695943 RGD:8695944 RGD:9479164
NCBI chr17:48,976,510...49,083,454
G
B
TET3
tet methylcytosine dioxygenase 3
ISO
RGD PMID:22770212 PMID:23697932 PMID:24825349 RGD:8695943 RGD:8695944 RGD:9479164
NCBI chr2A:74,058,948...74,180,046
G
C
Tet3
tet methylcytosine dioxygenase 3
ISO
RGD PMID:22770212 PMID:23697932 PMID:24825349 RGD:8695943 RGD:8695944 RGD:9479164
NCBI chrNW_004955424:11,612,124...11,722,130
G
R
Tet3
tet methylcytosine dioxygenase 3
ISO
RGD
PMID:22770212 PMID:23697932 PMID:24825349 RGD:8695943 , RGD:9479164 , RGD:8695944
NCBI chr 4:117,425,106...117,525,495
G
M
Tet3
tet methylcytosine dioxygenase 3
ISO
RGD
PMID:22770212 PMID:23697932 PMID:24825349 RGD:8695943 , RGD:9479164 , RGD:8695944
NCBI chr 6:83,339,355...83,434,190
G
H
TET3
tet methylcytosine dioxygenase 3
TAS
RGD
PMID:22770212 PMID:24825349 PMID:23697932 RGD:8695943 , RGD:8695944 , RGD:9479164
NCBI chr 2:73,983,631...74,135,498
G
B
Uhrf1
ubiquitin like with PHD and ring finger domains 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 4:87,693,066...87,712,181
G
N
Uhrf1
ubiquitin like with PHD and ring finger domains 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004624828:4,898,786...4,929,136
G
G
UHRF1
ubiquitin like with PHD and ring finger domains 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 6:4,592,362...4,642,179
G
P
UHRF1
ubiquitin like with PHD and ring finger domains 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 2:73,898,061...73,937,727
G
S
Uhrf1
ubiquitin like with PHD and ring finger domains 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004936588:2,863,651...2,890,116
G
D
UHRF1
ubiquitin like with PHD and ring finger domains 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr20:54,858,673...54,889,769
G
B
UHRF1
ubiquitin like with PHD and ring finger domains 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr19:3,926,583...3,979,163
G
C
Uhrf1
ubiquitin like with PHD and ring finger domains 1
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004955495:4,153,898...4,154,611
G
R
Uhrf1
ubiquitin-like with PHD and ring finger domains 1
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr 9:1,220,162...1,241,792
G
M
Uhrf1
ubiquitin-like, containing PHD and RING finger domains, 1
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr17:56,610,405...56,630,486
G
H
UHRF1
ubiquitin like with PHD and ring finger domains 1
TAS
RGD
PMID:23324617 RGD:8695954
NCBI chr19:4,903,080...4,962,154
G
B
Uhrf2
ubiquitin like with PHD and ring finger domains 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 2:30,136,382...30,199,794
G
N
Uhrf2
ubiquitin like with PHD and ring finger domains 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004624736:10,901,260...11,004,832
G
G
UHRF2
ubiquitin like with PHD and ring finger domains 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr12:73,010,697...73,106,163
G
P
UHRF2
ubiquitin like with PHD and ring finger domains 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 1:215,730,212...215,807,630
G
S
Uhrf2
ubiquitin like with PHD and ring finger domains 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004936539:910,191...990,258
G
D
UHRF2
ubiquitin like with PHD and ring finger domains 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr11:27,406,275...27,474,971
G
B
UHRF2
ubiquitin like with PHD and ring finger domains 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 9:6,228,895...6,322,319
G
C
Uhrf2
ubiquitin like with PHD and ring finger domains 2
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004955434:10,337,871...10,413,290
G
R
Uhrf2
ubiquitin like with PHD and ring finger domains 2
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr 1:237,224,050...237,291,406
G
M
Uhrf2
ubiquitin-like, containing PHD and RING finger domains 2
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr19:30,007,920...30,071,126
G
H
UHRF2
ubiquitin like with PHD and ring finger domains 2
TAS
RGD
PMID:23324617 RGD:8695954
NCBI chr 9:6,413,199...6,507,054
G
B
Zbtb33
zinc finger and BTB domain containing 33
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr X:101,521,516...101,528,686
G
N
Zbtb33
zinc finger and BTB domain containing 33
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004624931:61,085...69,372
G
G
ZBTB33
zinc finger and BTB domain containing 33
ISO
RGD PMID:23324617 RGD:8695954
G
P
ZBTB33
zinc finger and BTB domain containing 33
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr X:98,370,137...98,413,686
G
S
Zbtb33
zinc finger and BTB domain containing 33
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004936479:9,781,923...9,791,000
G
D
ZBTB33
zinc finger and BTB domain containing 33
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr X:92,243,159...92,250,940
G
B
ZBTB33
zinc finger and BTB domain containing 33
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr X:109,521,333...109,530,467
G
C
Zbtb33
zinc finger and BTB domain containing 33
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004955572:1,628,263...1,636,299
G
R
Zbtb33
zinc finger and BTB domain containing 33
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr X:121,828,960...121,836,193
G
M
Zbtb33
zinc finger and BTB domain containing 33
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr X:37,278,670...37,285,923
G
H
ZBTB33
zinc finger and BTB domain containing 33
TAS
RGD
PMID:23324617 RGD:8695954
NCBI chr X:120,250,812...120,258,398
G
B
Zbtb38
zinc finger and BTB domain containing 38
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 8:88,136,702...88,190,055
G
N
Zbtb38
zinc finger and BTB domain containing 38
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004624730:17,952,625...18,075,245
G
G
ZBTB38
zinc finger and BTB domain containing 38
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr15:49,160,877...49,203,378
G
P
ZBTB38
zinc finger and BTB domain containing 38
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr13:82,053,887...82,195,321
G
S
Zbtb38
zinc finger and BTB domain containing 38
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004936540:3,207,044...3,263,373
G
D
ZBTB38
zinc finger and BTB domain containing 38
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr23:37,283,794...37,344,316
G
B
ZBTB38
zinc finger and BTB domain containing 38
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 3:138,401,279...138,481,969
G
C
Zbtb38
zinc finger and BTB domain containing 38
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004955508:4,260,716...4,297,435
G
R
Zbtb38
zinc finger and BTB domain containing 38
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr 8:106,124,181...106,231,215
G
M
Zbtb38
zinc finger and BTB domain containing 38
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr 9:96,564,820...96,613,728
G
H
ZBTB38
zinc finger and BTB domain containing 38
TAS
RGD
PMID:23324617 RGD:8695954
NCBI chr 3:141,324,186...141,449,792
G
B
Zbtb4
zinc finger and BTB domain containing 4
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 9:49,363,778...49,387,286
G
N
Zbtb4
zinc finger and BTB domain containing 4
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004624786:10,067,503...10,085,500
G
G
ZBTB4
zinc finger and BTB domain containing 4
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr16:6,836,070...6,855,523
G
P
ZBTB4
zinc finger and BTB domain containing 4
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr12:52,759,912...52,775,725
G
S
Zbtb4
zinc finger and BTB domain containing 4
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004936595:736,873...753,386
G
D
ZBTB4
zinc finger and BTB domain containing 4
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 5:32,384,353...32,401,658
G
B
ZBTB4
zinc finger and BTB domain containing 4
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr17:7,491,971...7,516,823
G
C
Zbtb4
zinc finger and BTB domain containing 4
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004955467:9,452,423...9,470,494
G
R
Zbtb4
zinc finger and BTB domain containing 4
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr10:54,979,421...55,000,215
G
M
Zbtb4
zinc finger and BTB domain containing 4
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr11:69,656,738...69,674,852
G
H
ZBTB4
zinc finger and BTB domain containing 4
TAS
RGD
PMID:23324617 RGD:8695954
NCBI chr17:7,459,378...7,484,249
G
B
Zfp57
ZFP57 zinc finger protein
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr18:10,000,286...10,006,727
G
N
Zfp57
ZFP57 zinc finger protein
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004624754:25,390,107...25,397,205
G
G
ZFP57
ZFP57 zinc finger protein
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr17:42,942,002...42,947,064
G
P
ZFP57
ZFP57 zinc finger protein
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr 7:22,607,420...22,612,064
G
S
Zfp57
ZFP57 zinc finger protein
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004936723:1,063,969...1,069,986
G
D
ZFP57
ZFP57 zinc finger protein
ISO
RGD PMID:23324617 RGD:8695954
NCBI chr35:26,267,358...26,269,429
G
C
Zfp57
ZFP57 zinc finger protein
ISO
RGD PMID:23324617 RGD:8695954
NCBI chrNW_004955583:151,338...154,827
G
R
Zfp57
zinc finger protein 57
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr20:1,526,565...1,546,480
G
M
Zfp57
zinc finger protein 57
ISO
RGD
PMID:23324617 RGD:8695954
NCBI chr17:37,307,060...37,326,196
G
H
ZFP57
ZFP57 zinc finger protein
TAS
RGD
PMID:23324617 RGD:8695954
NCBI chr 6:29,672,392...29,681,152
G
B
Mbd4
methyl-CpG binding domain 4, DNA glycosylase
ISO
in silico assessment of loss of DNA-protein interactions and observed enhancement of DNA repair capacity (DRC) in the context of nucleotide excision repair (NER) pathway for SNP
RGD PMID:24004570 RGD:9588971
NCBI chr 6:30,872,004...30,882,213
G
N
Mbd4
methyl-CpG binding domain 4, DNA glycosylase
ISO
in silico assessment of loss of DNA-protein interactions and observed enhancement of DNA repair capacity (DRC) in the context of nucleotide excision repair (NER) pathway for SNP
RGD PMID:24004570 RGD:9588971
NCBI chrNW_004624872:453,303...462,661
G
G
MBD4
methyl-CpG binding domain 4, DNA glycosylase
ISO
in silico assessment of loss of DNA-protein interactions and observed enhancement of DNA repair capacity (DRC) in the context of nucleotide excision repair (NER) pathway for SNP
RGD PMID:24004570 RGD:9588971
NCBI chr22:51,616,840...51,626,232
G
P
MBD4
methyl-CpG binding domain 4, DNA glycosylase
ISO
in silico assessment of loss of DNA-protein interactions and observed enhancement of DNA repair capacity (DRC) in the context of nucleotide excision repair (NER) pathway for SNP
RGD PMID:24004570 RGD:9588971
NCBI chr13:68,816,677...68,828,559
G
S
Mbd4
methyl-CpG binding domain 4, DNA glycosylase
ISO
in silico assessment of loss of DNA-protein interactions and observed enhancement of DNA repair capacity (DRC) in the context of nucleotide excision repair (NER) pathway for SNP
RGD PMID:24004570 RGD:9588971
NCBI chrNW_004936602:968,159...980,452
G
D
MBD4
methyl-CpG binding domain 4, DNA glycosylase
ISO
in silico assessment of loss of DNA-protein interactions and observed enhancement of DNA repair capacity (DRC) in the context of nucleotide excision repair (NER) pathway for SNP
RGD PMID:24004570 RGD:9588971
NCBI chr20:5,716,162...5,725,068
G
B
MBD4
methyl-CpG binding domain 4, DNA glycosylase
ISO
in silico assessment of loss of DNA-protein interactions and observed enhancement of DNA repair capacity (DRC) in the context of nucleotide excision repair (NER) pathway for SNP
RGD PMID:24004570 RGD:9588971
NCBI chr 3:126,489,125...126,498,034
G
C
Mbd4
methyl-CpG binding domain 4, DNA glycosylase
ISO
in silico assessment of loss of DNA-protein interactions and observed enhancement of DNA repair capacity (DRC) in the context of nucleotide excision repair (NER) pathway for SNP
RGD PMID:24004570 RGD:9588971
NCBI chrNW_004955429:17,858,656...17,868,111
G
R
Mbd4
methyl-CpG binding domain 4 DNA glycosylase
ISO
in silico assessment of loss of DNA-protein interactions and observed enhancement of DNA repair capacity (DRC) in the context of nucleotide excision repair (NER) pathway for SNP
RGD
PMID:24004570 RGD:9588971
NCBI chr 4:150,565,646...150,577,433
G
M
Mbd4
methyl-CpG binding domain protein 4
ISO
in silico assessment of loss of DNA-protein interactions and observed enhancement of DNA repair capacity (DRC) in the context of nucleotide excision repair (NER) pathway for SNP
RGD
PMID:24004570 RGD:9588971
NCBI chr 6:115,817,658...115,830,361
G
H
MBD4
methyl-CpG binding domain 4, DNA glycosylase
IMP
in silico assessment of loss of DNA-protein interactions and observed enhancement of DNA repair capacity (DRC) in the context of nucleotide excision repair (NER) pathway for SNP
RGD
PMID:24004570 RGD:9588971
NCBI chr 3:129,430,947...129,439,948
Pathway Gene Annotations
Disease Annotations Associated with Genes in the DNA modification pathway
Dnmt1 acute myeloid leukemia , adenocarcinoma , alopecia areata , anxiety disorder , asthma , Ataxia , autism spectrum disorder , autoimmune disease , autosomal dominant cerebellar ataxia, deafness and narcolepsy , Beckwith-Wiedemann syndrome , Binge Drinking , Brain Injuries , Breast Neoplasms , bronchiolo-alveolar adenocarcinoma , Carcinogenesis , carcinoma , cardiomyopathy , cerebellar ataxia , Charcot-Marie-Tooth disease , choline deficiency disease , chronic myeloid leukemia , clear cell renal cell carcinoma , Colonic Neoplasms , Colorectal Neoplasms , congenital heart disease , congestive heart failure , dementia , Endometrial Neoplasms , Endometrioid Carcinomas , endometriosis , endometriosis of uterus , Experimental Arthritis , fetal alcohol spectrum disorder , Fetal Growth Retardation , genetic disease , Graves' disease , Hearing Loss , hepatocellular carcinoma , hereditary sensory neuropathy , hereditary sensory neuropathy type 1E , hypertrophic cardiomyopathy , liver cirrhosis , lung cancer , lung carcinoma , lung non-small cell carcinoma , medulloblastoma , metabolic dysfunction and alcohol associated liver disease , middle cerebral artery infarction , mucoepidermoid carcinoma , myelodysplastic syndrome , Neoplasm Invasiveness , Oral Lichen Planus , Ovarian Neoplasms , pancreatic ductal carcinoma , Pancreatic Intraepithelial Neoplasia , Peritoneal Fibrosis , Pituitary Stalk Interruption Syndrome , prostate adenocarcinoma , prostate cancer , prostate carcinoma in situ , Prostatic Neoplasms , rheumatoid arthritis , schizophrenia , spastic ataxia , Spastic Paraparesis , Stomach Neoplasms , systemic lupus erythematosus , type 2 diabetes mellitus , urinary bladder cancer , Uterine Cervical Neoplasms DNMT1 acute myeloid leukemia , adenocarcinoma , alopecia areata , anxiety disorder , asthma , Ataxia , autism spectrum disorder , autoimmune disease , autosomal dominant cerebellar ataxia, deafness and narcolepsy , Beckwith-Wiedemann syndrome , Binge Drinking , Brain Injuries , Breast Neoplasms , bronchiolo-alveolar adenocarcinoma , Carcinogenesis , carcinoma , cardiomyopathy , cerebellar ataxia , Charcot-Marie-Tooth disease , choline deficiency disease , chronic myeloid leukemia , clear cell renal cell carcinoma , Colonic Neoplasms , Colorectal Neoplasms , congenital heart disease , congestive heart failure , dementia , Endometrial Neoplasms , Endometrioid Carcinomas , endometriosis , endometriosis of uterus , Experimental Arthritis , fetal alcohol spectrum disorder , Fetal Growth Retardation , genetic disease , Graves' disease , Hearing Loss , hepatocellular carcinoma , hereditary sensory neuropathy , hereditary sensory neuropathy type 1E , hypertrophic cardiomyopathy , liver cirrhosis , lung cancer , lung carcinoma , lung non-small cell carcinoma , medulloblastoma , metabolic dysfunction and alcohol associated liver disease , middle cerebral artery infarction , mucoepidermoid carcinoma , myelodysplastic syndrome , Neoplasm Invasiveness , Oral Lichen Planus , Ovarian Neoplasms , pancreatic ductal carcinoma , Pancreatic Intraepithelial Neoplasia , Peritoneal Fibrosis , Pituitary Stalk Interruption Syndrome , prostate adenocarcinoma , prostate cancer , prostate carcinoma in situ , Prostatic Neoplasms , rheumatoid arthritis , schizophrenia , spastic ataxia , Spastic Paraparesis , Stomach Neoplasms , systemic lupus erythematosus , type 2 diabetes mellitus , urinary bladder cancer , Uterine Cervical Neoplasms Dnmt1 acute myeloid leukemia , adenocarcinoma , alopecia areata , anxiety disorder , asthma , Ataxia , autism spectrum disorder , autoimmune disease , autosomal dominant cerebellar ataxia, deafness and narcolepsy , Beckwith-Wiedemann syndrome , Binge Drinking , Brain Injuries , Breast Neoplasms , bronchiolo-alveolar adenocarcinoma , Carcinogenesis , carcinoma , cardiomyopathy , cerebellar ataxia , Charcot-Marie-Tooth disease , choline deficiency disease , chronic myeloid leukemia , clear cell renal cell carcinoma , Colonic Neoplasms , Colorectal Neoplasms , congenital heart disease , congestive heart failure , dementia , Endometrial Neoplasms , Endometrioid Carcinomas , endometriosis , endometriosis of uterus , Experimental Arthritis , fetal alcohol spectrum disorder , Fetal Growth Retardation , genetic disease , Graves' disease , Hearing Loss , hepatocellular carcinoma , hereditary sensory neuropathy , hereditary sensory neuropathy type 1E , hypertrophic cardiomyopathy , liver cirrhosis , lung cancer , lung carcinoma , lung non-small cell carcinoma , medulloblastoma , metabolic dysfunction and alcohol associated liver disease , middle cerebral artery infarction , mucoepidermoid carcinoma , myelodysplastic syndrome , Neoplasm Invasiveness , Oral Lichen Planus , Ovarian Neoplasms , pancreatic ductal carcinoma , Pancreatic Intraepithelial Neoplasia , Peritoneal Fibrosis , Pituitary Stalk Interruption Syndrome , prostate adenocarcinoma , prostate cancer , prostate carcinoma in situ , Prostatic Neoplasms , rheumatoid arthritis , schizophrenia , spastic ataxia , Spastic Paraparesis , Stomach Neoplasms , systemic lupus erythematosus , type 2 diabetes mellitus , urinary bladder cancer , Uterine Cervical Neoplasms Dnmt3a acute lymphoblastic leukemia , acute monocytic leukemia , acute myeloid leukemia , acute promyelocytic leukemia , amyotrophic lateral sclerosis , autism spectrum disorder , autoimmune thrombocytopenic purpura , bone marrow disease , Brain Hypoxia-Ischemia , Breast Neoplasms , Carcinogenesis , chronic myeloid leukemia , clear cell renal cell carcinoma , congenital heart disease , Craniofacial Abnormalities , Crohn's disease , depressive disorder , Desbuquois dysplasia , diffuse large B-cell lymphoma , Dwarfism , endometriosis , endometriosis of uterus , Facies , genetic disease , glioblastoma , Growth Disorders , hepatocellular carcinoma , Heyn-Sproul-Jackson Syndrome , Immunoblastic Lymphadenopathy , intellectual disability , juvenile myelomonocytic leukemia , lung adenocarcinoma , lung cancer , lung carcinoma , Lung Neoplasms , melanoma , microcephaly , middle cerebral artery infarction , multiple myeloma , Muscle Hypotonia , myelodysplastic syndrome , myeloid neoplasm , Neoplasm Recurrence, Local , Nervous System Malformations , Neurodevelopmental Disorders , obesity , peripheral T-cell lymphoma , primary cutaneous T-cell non-Hodgkin lymphoma , prostate adenocarcinoma , prostatitis , sciatic neuropathy , Sezary's disease , stomach cancer , T-cell acute lymphoblastic leukemia , Tatton-Brown-Rahman syndrome DNMT3A acute lymphoblastic leukemia , acute monocytic leukemia , acute myeloid leukemia , acute promyelocytic leukemia , amyotrophic lateral sclerosis , autism spectrum disorder , autoimmune thrombocytopenic purpura , bone marrow disease , Brain Hypoxia-Ischemia , Breast Neoplasms , Carcinogenesis , chronic myeloid leukemia , clear cell renal cell carcinoma , congenital heart disease , Craniofacial Abnormalities , Crohn's disease , depressive disorder , Desbuquois dysplasia , diffuse large B-cell lymphoma , Dwarfism , endometriosis , endometriosis of uterus , Facies , genetic disease , glioblastoma , Growth Disorders , hepatocellular carcinoma , Heyn-Sproul-Jackson Syndrome , Immunoblastic Lymphadenopathy , intellectual disability , juvenile myelomonocytic leukemia , lung cancer , lung carcinoma , Lung Neoplasms , melanoma , microcephaly , middle cerebral artery infarction , multiple myeloma , Muscle Hypotonia , myelodysplastic syndrome , myeloid neoplasm , Neoplasm Recurrence, Local , Nervous System Malformations , Neurodevelopmental Disorders , obesity , peripheral T-cell lymphoma , primary cutaneous T-cell non-Hodgkin lymphoma , prostate adenocarcinoma , prostatitis , sciatic neuropathy , Sezary's disease , stomach cancer , T-cell acute lymphoblastic leukemia , Tatton-Brown-Rahman syndrome Dnmt3a acute lymphoblastic leukemia , acute monocytic leukemia , acute myeloid leukemia , acute promyelocytic leukemia , amyotrophic lateral sclerosis , autism spectrum disorder , autoimmune thrombocytopenic purpura , bone marrow disease , Brain Hypoxia-Ischemia , Breast Neoplasms , Carcinogenesis , chronic myeloid leukemia , clear cell renal cell carcinoma , congenital heart disease , Craniofacial Abnormalities , Crohn's disease , depressive disorder , Desbuquois dysplasia , diffuse large B-cell lymphoma , Dwarfism , endometriosis , endometriosis of uterus , Facies , genetic disease , glioblastoma , Growth Disorders , hepatocellular carcinoma , Heyn-Sproul-Jackson Syndrome , Immunoblastic Lymphadenopathy , intellectual disability , juvenile myelomonocytic leukemia , lung adenocarcinoma , lung cancer , lung carcinoma , Lung Neoplasms , melanoma , microcephaly , middle cerebral artery infarction , multiple myeloma , Muscle Hypotonia , myelodysplastic syndrome , myeloid neoplasm , Neoplasm Recurrence, Local , Nervous System Malformations , Neurodevelopmental Disorders , obesity , peripheral T-cell lymphoma , primary cutaneous T-cell non-Hodgkin lymphoma , prostate adenocarcinoma , prostatitis , sciatic neuropathy , Sezary's disease , stomach cancer , T-cell acute lymphoblastic leukemia , Tatton-Brown-Rahman syndrome Dnmt3b acute myeloid leukemia , anxiety disorder , autism spectrum disorder , autoimmune thrombocytopenic purpura , B-Cell Chronic Lymphocytic Leukemia , Breast Neoplasms , carcinoma , chronic myeloid leukemia , colorectal adenocarcinoma , Colorectal Neoplasms , congenital heart disease , Craniofacial Abnormalities , Dysmenorrhea , Endometrial Neoplasms , Endometrioid Carcinomas , endometriosis , endometriosis of uterus , esophagus squamous cell carcinoma , Experimental Mammary Neoplasms , facioscapulohumeral muscular dystrophy 4 , Familial Prostate Cancer , Female Infertility , fetal alcohol spectrum disorder , Fetal Growth Retardation , gastric body carcinoma , genetic disease , head and neck squamous cell carcinoma , hepatocellular carcinoma , immunodeficiency-centromeric instability-facial anomalies syndrome , immunodeficiency-centromeric instability-facial anomalies syndrome 1 , Kabuki Syndrome 1 , lung adenocarcinoma , lung cancer , lung small cell carcinoma , Lymphatic Metastasis , Multiple Primary Neoplasms , myelodysplastic syndrome , Neoplasm Metastasis , Oral Lichen Planus , oral squamous cell carcinoma , pancreatic ductal carcinoma , primary immunodeficiency disease , prostate adenocarcinoma , Prostatic Neoplasms , prostatitis , rheumatoid arthritis , schizophrenia , skin melanoma , Stomach Neoplasms , thymoma , type 1 diabetes mellitus , Uterine Cervical Neoplasms Dnmt3b acute myeloid leukemia , anxiety disorder , autism spectrum disorder , autoimmune thrombocytopenic purpura , B-Cell Chronic Lymphocytic Leukemia , Breast Neoplasms , carcinoma , chronic myeloid leukemia , colorectal adenocarcinoma , Colorectal Neoplasms , congenital heart disease , Craniofacial Abnormalities , Dysmenorrhea , Endometrial Neoplasms , Endometrioid Carcinomas , endometriosis , endometriosis of uterus , esophagus squamous cell carcinoma , Experimental Mammary Neoplasms , facioscapulohumeral muscular dystrophy 4 , Familial Prostate Cancer , Female Infertility , fetal alcohol spectrum disorder , Fetal Growth Retardation , gastric body carcinoma , genetic disease , head and neck squamous cell carcinoma , hepatocellular carcinoma , immunodeficiency-centromeric instability-facial anomalies syndrome , immunodeficiency-centromeric instability-facial anomalies syndrome 1 , Kabuki Syndrome 1 , lung adenocarcinoma , lung cancer , lung small cell carcinoma , Lymphatic Metastasis , Multiple Primary Neoplasms , myelodysplastic syndrome , Neoplasm Metastasis , Oral Lichen Planus , oral squamous cell carcinoma , pancreatic ductal carcinoma , primary immunodeficiency disease , prostate adenocarcinoma , Prostatic Neoplasms , prostatitis , rheumatoid arthritis , schizophrenia , skin melanoma , Stomach Neoplasms , thymoma , type 1 diabetes mellitus , Uterine Cervical Neoplasms DNMT3B acute myeloid leukemia , anxiety disorder , autism spectrum disorder , autoimmune thrombocytopenic purpura , B-Cell Chronic Lymphocytic Leukemia , Breast Neoplasms , carcinoma , chronic myeloid leukemia , colorectal adenocarcinoma , Colorectal Neoplasms , congenital heart disease , Craniofacial Abnormalities , Dysmenorrhea , Endometrial Neoplasms , Endometrioid Carcinomas , endometriosis , endometriosis of uterus , esophagus squamous cell carcinoma , Experimental Mammary Neoplasms , facioscapulohumeral muscular dystrophy 4 , Familial Prostate Cancer , Female Infertility , fetal alcohol spectrum disorder , Fetal Growth Retardation , gastric body carcinoma , genetic disease , head and neck squamous cell carcinoma , hepatocellular carcinoma , immunodeficiency-centromeric instability-facial anomalies syndrome , immunodeficiency-centromeric instability-facial anomalies syndrome 1 , Kabuki Syndrome 1 , lung adenocarcinoma , lung cancer , lung small cell carcinoma , Lymphatic Metastasis , Multiple Primary Neoplasms , myelodysplastic syndrome , Neoplasm Metastasis , Oral Lichen Planus , oral squamous cell carcinoma , pancreatic ductal carcinoma , primary immunodeficiency disease , prostate adenocarcinoma , Prostatic Neoplasms , prostatitis , rheumatoid arthritis , schizophrenia , skin melanoma , Stomach Neoplasms , thymoma , type 1 diabetes mellitus , Uterine Cervical Neoplasms Ebf1 adult respiratory distress syndrome , follicular lymphoma , lipodystrophy , Neoplastic Cell Transformation Ebf1 adult respiratory distress syndrome , follicular lymphoma , lipodystrophy , Neoplastic Cell Transformation EBF1 adult respiratory distress syndrome , follicular lymphoma , lipodystrophy , Neoplastic Cell Transformation Kdm2a Colorectal Neoplasms , congestive heart failure , Familial Prostate Cancer , lung non-small cell carcinoma , Neurodevelopmental Disorders KDM2A Colorectal Neoplasms , congestive heart failure , Familial Prostate Cancer , lung non-small cell carcinoma , Neurodevelopmental Disorders Kdm2a Colorectal Neoplasms , congestive heart failure , Familial Prostate Cancer , lung non-small cell carcinoma , Neurodevelopmental Disorders Kdm2b acute lymphoblastic leukemia , Developmental Disabilities , genetic disease , neural tube defect , Neurodevelopmental Disorders , pancreatic ductal carcinoma , schizophrenia , severe combined immunodeficiency , T-cell non-Hodgkin lymphoma KDM2B acute lymphoblastic leukemia , Developmental Disabilities , genetic disease , neural tube defect , Neurodevelopmental Disorders , pancreatic ductal carcinoma , schizophrenia , severe combined immunodeficiency , T-cell non-Hodgkin lymphoma Kdm2b acute lymphoblastic leukemia , Developmental Disabilities , genetic disease , neural tube defect , Neurodevelopmental Disorders , pancreatic ductal carcinoma , schizophrenia , severe combined immunodeficiency , T-cell non-Hodgkin lymphoma Kmt2a Acute Erythroleukemia , acute lymphoblastic leukemia , acute monocytic leukemia , acute myeloid leukemia , acute myelomonocytic leukemia , adenocarcinoma , autism spectrum disorder , cervical cancer , Chromosome Aberrations , Cornelia de Lange syndrome 1 , Desbuquois dysplasia , Developmental Disease , genetic disease , hepatocellular carcinoma , Hirsutism , intellectual disability , Kabuki Syndrome 1 , Kidney Reperfusion Injury , language disorder , leukemia , lung small cell carcinoma , lymphoid leukemia , microcephaly , myelofibrosis , myeloid leukemia , Neurodevelopmental Disorders , non-Hodgkin lymphoma , prostate cancer , Prostatic Neoplasms , Rubinstein Taybi like Syndrome , Stomach Neoplasms , tetralogy of Fallot , transient cerebral ischemia , transitional cell carcinoma , urinary bladder cancer , Wiedemann-Steiner syndrome KMT2A Acute Erythroleukemia , acute lymphoblastic leukemia , acute monocytic leukemia , acute myeloid leukemia , acute myelomonocytic leukemia , adenocarcinoma , autism spectrum disorder , cervical cancer , Chromosome Aberrations , Cornelia de Lange syndrome 1 , Desbuquois dysplasia , Developmental Disease , genetic disease , hepatocellular carcinoma , Hirsutism , intellectual disability , Kabuki Syndrome 1 , Kidney Reperfusion Injury , language disorder , leukemia , lung small cell carcinoma , lymphoid leukemia , microcephaly , myelofibrosis , myeloid leukemia , Neurodevelopmental Disorders , non-Hodgkin lymphoma , prostate cancer , Prostatic Neoplasms , Rubinstein Taybi like Syndrome , Stomach Neoplasms , tetralogy of Fallot , transient cerebral ischemia , transitional cell carcinoma , urinary bladder cancer , Wiedemann-Steiner syndrome Kmt2a Acute Erythroleukemia , acute lymphoblastic leukemia , acute monocytic leukemia , acute myeloid leukemia , acute myelomonocytic leukemia , adenocarcinoma , autism spectrum disorder , cervical cancer , Chromosome Aberrations , Cornelia de Lange syndrome 1 , Desbuquois dysplasia , Developmental Disease , genetic disease , hepatocellular carcinoma , Hirsutism , intellectual disability , Kabuki Syndrome 1 , Kidney Reperfusion Injury , language disorder , leukemia , lung small cell carcinoma , lymphoid leukemia , microcephaly , myelofibrosis , myeloid leukemia , Neurodevelopmental Disorders , non-Hodgkin lymphoma , prostate cancer , Prostatic Neoplasms , Rubinstein Taybi like Syndrome , Stomach Neoplasms , tetralogy of Fallot , transient cerebral ischemia , transitional cell carcinoma , urinary bladder cancer , Wiedemann-Steiner syndrome KMT2C acute myeloid leukemia , adenocarcinoma , adenoid cystic carcinoma , autism spectrum disorder , cholangiocarcinoma , developmental and epileptic encephalopathy 92 , Developmental Disabilities , esophagus squamous cell carcinoma , genetic disease , hepatocellular carcinoma , intellectual disability , Klatskin's tumor , Kleefstra syndrome 1 , Kleefstra syndrome 2 , laryngeal carcinoma , lung adenocarcinoma , lung non-small cell carcinoma , microcephaly , multiple myeloma , nasopharynx carcinoma , Neurodevelopmental Disorders , Parasitic Liver Diseases , prostate cancer , Prostatic Neoplasms , rheumatoid arthritis , Sezary's disease , squamous cell carcinoma , stomach cancer , stomach carcinoma , Stomach Neoplasms , syndromic intellectual disability , transitional cell carcinoma , urinary bladder cancer Kmt2c acute myeloid leukemia , adenocarcinoma , adenoid cystic carcinoma , autism spectrum disorder , cholangiocarcinoma , developmental and epileptic encephalopathy 92 , Developmental Disabilities , esophagus squamous cell carcinoma , genetic disease , hepatocellular carcinoma , intellectual disability , Klatskin's tumor , Kleefstra syndrome 1 , Kleefstra syndrome 2 , laryngeal carcinoma , lung adenocarcinoma , lung non-small cell carcinoma , microcephaly , multiple myeloma , nasopharynx carcinoma , Neurodevelopmental Disorders , Parasitic Liver Diseases , prostate cancer , Prostatic Neoplasms , rheumatoid arthritis , Sezary's disease , squamous cell carcinoma , stomach cancer , stomach carcinoma , Stomach Neoplasms , syndromic intellectual disability , transitional cell carcinoma , urinary bladder cancer Kmt2c acute myeloid leukemia , adenocarcinoma , adenoid cystic carcinoma , autism spectrum disorder , cholangiocarcinoma , developmental and epileptic encephalopathy 92 , Developmental Disabilities , esophagus squamous cell carcinoma , genetic disease , hepatocellular carcinoma , intellectual disability , Klatskin's tumor , Kleefstra syndrome 1 , Kleefstra syndrome 2 , laryngeal carcinoma , lung adenocarcinoma , lung non-small cell carcinoma , microcephaly , multiple myeloma , nasopharynx carcinoma , Neurodevelopmental Disorders , Parasitic Liver Diseases , prostate cancer , Prostatic Neoplasms , rheumatoid arthritis , Sezary's disease , squamous cell carcinoma , stomach cancer , stomach carcinoma , Stomach Neoplasms , syndromic intellectual disability , transitional cell carcinoma , urinary bladder cancer Mbd1 autistic disorder , Experimental Seizures , lung cancer , Prostatic Neoplasms , transient cerebral ischemia MBD1 autistic disorder , Experimental Seizures , lung cancer , Prostatic Neoplasms , transient cerebral ischemia Mbd1 autistic disorder , Experimental Seizures , lung cancer , Prostatic Neoplasms , transient cerebral ischemia MBD2 Brain Hypoxia-Ischemia , choline deficiency disease , Experimental Seizures , Familial Prostate Cancer , macular degeneration , Prostatic Neoplasms , rheumatoid arthritis , schizophrenia , transient cerebral ischemia , type 1 diabetes mellitus Mbd2 Brain Hypoxia-Ischemia , choline deficiency disease , Experimental Seizures , Familial Prostate Cancer , macular degeneration , Prostatic Neoplasms , rheumatoid arthritis , schizophrenia , transient cerebral ischemia , type 1 diabetes mellitus Mbd2 Brain Hypoxia-Ischemia , choline deficiency disease , Experimental Seizures , Familial Prostate Cancer , macular degeneration , Prostatic Neoplasms , rheumatoid arthritis , schizophrenia , transient cerebral ischemia , type 1 diabetes mellitus MBD4 autistic disorder , colorectal carcinoma , Colorectal Neoplasms , esophagus squamous cell carcinoma , genetic disease , Hereditary Neoplastic Syndromes , Intestinal Neoplasms , lung cancer , rheumatoid arthritis , Tumor Predisposition Syndrome 2 , uveal melanoma Mbd4 autistic disorder , colorectal carcinoma , Colorectal Neoplasms , esophagus squamous cell carcinoma , genetic disease , Hereditary Neoplastic Syndromes , Intestinal Neoplasms , lung cancer , rheumatoid arthritis , Tumor Predisposition Syndrome 2 , uveal melanoma Mbd4 autistic disorder , colorectal carcinoma , Colorectal Neoplasms , esophagus squamous cell carcinoma , genetic disease , Hereditary Neoplastic Syndromes , Intestinal Neoplasms , lung cancer , rheumatoid arthritis , Tumor Predisposition Syndrome 2 , uveal melanoma Mecp2 Angelman syndrome , Animal Disease Models , anxiety disorder , attention deficit hyperactivity disorder , autism spectrum disorder , autistic disorder , Binge Drinking , Cocaine-Related Disorders , congenital variant of Rett syndrome , Craniofacial Abnormalities , developmental and epileptic encephalopathy 2 , Developmental Disabilities , Developmental Disease , epilepsy , Experimental Seizures , Facial Hypertrichosis , fetal alcohol spectrum disorder , Fetal Growth Retardation , focal epilepsy , gastrointestinal system disease , generalized epilepsy , genetic disease , Hearing Loss , hepatocellular carcinoma , Hyperalgesia , intellectual disability , learning disability , microcephaly , Micrognathism , Muscle Hypotonia , Myoclonus , Nervous System Malformations , Neurodevelopmental Disorders , non-syndromic X-linked intellectual disability , periventricular nodular heterotopia , pervasive developmental disorder , Psychomotor Disorders , pulmonary fibrosis , respiratory failure , Rett syndrome , Rett Syndrome, Atypical , Rett Syndrome, Zappella Variant , schizophrenia , severe congenital encephalopathy due to MECP2 mutation , Smith-Magenis syndrome , stereotypic movement disorder , syndromic X-linked intellectual disability Lubs type , transient cerebral ischemia , X-Linked Intellectual Developmental Disorders , X-linked intellectual disability-psychosis-macroorchidism syndrome Mecp2 Angelman syndrome , Animal Disease Models , anxiety disorder , attention deficit hyperactivity disorder , autism spectrum disorder , autistic disorder , Binge Drinking , Cocaine-Related Disorders , congenital variant of Rett syndrome , Craniofacial Abnormalities , developmental and epileptic encephalopathy 2 , Developmental Disabilities , Developmental Disease , epilepsy , Experimental Seizures , Facial Hypertrichosis , fetal alcohol spectrum disorder , Fetal Growth Retardation , focal epilepsy , gastrointestinal system disease , generalized epilepsy , genetic disease , Hearing Loss , hepatocellular carcinoma , Hyperalgesia , intellectual disability , learning disability , microcephaly , Micrognathism , Muscle Hypotonia , Myoclonus , Nervous System Malformations , Neurodevelopmental Disorders , non-syndromic X-linked intellectual disability , periventricular nodular heterotopia , pervasive developmental disorder , Psychomotor Disorders , pulmonary fibrosis , respiratory failure , Rett syndrome , Rett Syndrome, Atypical , Rett Syndrome, Zappella Variant , schizophrenia , severe congenital encephalopathy due to MECP2 mutation , Smith-Magenis syndrome , stereotypic movement disorder , syndromic X-linked intellectual disability Lubs type , transient cerebral ischemia , X-Linked Intellectual Developmental Disorders , X-linked intellectual disability-psychosis-macroorchidism syndrome MECP2 Angelman syndrome , Animal Disease Models , anxiety disorder , attention deficit hyperactivity disorder , autism spectrum disorder , autistic disorder , Binge Drinking , Cocaine-Related Disorders , congenital variant of Rett syndrome , Craniofacial Abnormalities , developmental and epileptic encephalopathy 2 , Developmental Disabilities , Developmental Disease , epilepsy , Experimental Seizures , Facial Hypertrichosis , fetal alcohol spectrum disorder , Fetal Growth Retardation , focal epilepsy , gastrointestinal system disease , generalized epilepsy , genetic disease , Hearing Loss , hepatocellular carcinoma , Hyperalgesia , intellectual disability , learning disability , microcephaly , Micrognathism , Muscle Hypotonia , Myoclonus , Nervous System Malformations , Neurodevelopmental Disorders , non-syndromic X-linked intellectual disability , periventricular nodular heterotopia , pervasive developmental disorder , Psychomotor Disorders , pulmonary fibrosis , respiratory failure , Rett syndrome , Rett Syndrome, Atypical , Rett Syndrome, Zappella Variant , schizophrenia , severe congenital encephalopathy due to MECP2 mutation , Smith-Magenis syndrome , stereotypic movement disorder , syndromic X-linked intellectual disability Lubs type , transient cerebral ischemia , X-Linked Intellectual Developmental Disorders , X-linked intellectual disability-psychosis-macroorchidism syndrome Nanog colon cancer , Germ Cell and Embryonal Neoplasms , Neoplastic Cell Transformation , Parkinson's disease , teratoma NANOG colon cancer , Germ Cell and Embryonal Neoplasms , Neoplastic Cell Transformation , Parkinson's disease , teratoma Nanog colon cancer , Germ Cell and Embryonal Neoplasms , Neoplastic Cell Transformation , Parkinson's disease , teratoma Ogt aortic valve stenosis , Breast Neoplasms , Cardiomegaly , congenital disorder of glycosylation , congestive heart failure , Experimental Diabetes Mellitus , genetic disease , non-syndromic X-linked intellectual disability 106 Ogt aortic valve stenosis , Breast Neoplasms , Cardiomegaly , congenital disorder of glycosylation , congestive heart failure , Experimental Diabetes Mellitus , genetic disease , non-syndromic X-linked intellectual disability 106 OGT aortic valve stenosis , Breast Neoplasms , Cardiomegaly , congenital disorder of glycosylation , congestive heart failure , Experimental Diabetes Mellitus , genetic disease , non-syndromic X-linked intellectual disability 106 Pparg abdominal obesity-metabolic syndrome , acute kidney failure , Acute Lung Injury , acute promyelocytic leukemia , adenocarcinoma , alcoholic cardiomyopathy , Alveolar Bone Loss , Alzheimer's disease , antiphospholipid syndrome , arteriosclerosis , atherosclerosis , autosomal dominant polycystic kidney disease , Barrett's esophagus , cardiovascular system disease , carotid artery disease , Carotid Artery Injuries , Carotid Intimal Medial Thickness 1 , Chemical and Drug Induced Liver Injury , Chronic Allograft Nephropathy , Chronobiology Disorders , cognitive disorder , colon cancer , colon carcinoma , Colonic Neoplasms , Colorectal Neoplasms , congenital generalized lipodystrophy type 2 , congestive heart failure , corneal neovascularization , coronary artery disease , COVID-19 , Crohn's disease , diabetes mellitus , diabetic encephalopathy , Diabetic Nephropathies , diabetic retinopathy , disease of metabolism , Dyslipidemias , end stage renal disease , Endotoxemia , Experimental Autoimmune Myocarditis , Experimental Diabetes Mellitus , Experimental Liver Cirrhosis , Eye Burns , Eye Injuries , familial partial lipodystrophy , familial partial lipodystrophy type 3 , Fetal Growth Retardation , Fibrosis , Follicular Thyroid Cancer , genetic disease , glioblastoma , glomerulonephritis , glomerulosclerosis , Graves ophthalmopathy , hepatocellular carcinoma , high grade glioma , hyperandrogenism , hypertension , Inflammation , Insulin Resistance , ischemia , kidney failure , Left Ventricular Hypertrophy , leukostasis , lipid storage disease , lipodystrophy , Liver Neoplasms , Lymphatic Metastasis , maturity-onset diabetes of the young type 1 , melanoma , metabolic dysfunction-associated steatotic liver disease , Metabolic Syndrome , morbid obesity , Myocardial Reperfusion Injury , Nerve Degeneration , obesity , osteoarthritis , Oxygen-Induced Retinopathy , Pain , pancreatic cancer , pancreatic ductal carcinoma , pancreatitis , Perennial Allergic Rhinitis , pituitary-dependent Cushing's disease , polycystic kidney disease , pre-eclampsia , primary pulmonary hypertension , proliferative diabetic retinopathy , psoriasis , Reperfusion Injury , steatotic liver disease , Stomach Neoplasms , Subarachnoid Hemorrhage , systemic lupus erythematosus , T-cell non-Hodgkin lymphoma , Thyroid Neoplasms , transient cerebral ischemia , type 2 diabetes mellitus , urinary bladder cancer PPARG abdominal obesity-metabolic syndrome , acute kidney failure , Acute Lung Injury , acute promyelocytic leukemia , adenocarcinoma , alcoholic cardiomyopathy , Alveolar Bone Loss , Alzheimer's disease , antiphospholipid syndrome , arteriosclerosis , atherosclerosis , autosomal dominant polycystic kidney disease , Barrett's esophagus , cardiovascular system disease , carotid artery disease , Carotid Artery Injuries , Carotid Intimal Medial Thickness 1 , Chemical and Drug Induced Liver Injury , Chronic Allograft Nephropathy , Chronobiology Disorders , cognitive disorder , colon cancer , colon carcinoma , Colonic Neoplasms , Colorectal Neoplasms , congenital generalized lipodystrophy type 2 , congestive heart failure , corneal neovascularization , coronary artery disease , COVID-19 , Crohn's disease , diabetes mellitus , diabetic encephalopathy , Diabetic Nephropathies , diabetic retinopathy , disease of metabolism , Dyslipidemias , end stage renal disease , Endotoxemia , Experimental Autoimmune Myocarditis , Experimental Diabetes Mellitus , Experimental Liver Cirrhosis , Eye Burns , Eye Injuries , familial partial lipodystrophy , familial partial lipodystrophy type 3 , Fetal Growth Retardation , Fibrosis , Follicular Thyroid Cancer , genetic disease , glioblastoma , glomerulonephritis , glomerulosclerosis , Graves ophthalmopathy , hepatocellular carcinoma , high grade glioma , hyperandrogenism , hypertension , Inflammation , Insulin Resistance , ischemia , kidney failure , Left Ventricular Hypertrophy , leukostasis , lipid storage disease , lipodystrophy , Liver Neoplasms , Lymphatic Metastasis , maturity-onset diabetes of the young type 1 , melanoma , metabolic dysfunction-associated steatotic liver disease , Metabolic Syndrome , morbid obesity , Myocardial Reperfusion Injury , Nerve Degeneration , obesity , osteoarthritis , Oxygen-Induced Retinopathy , Pain , pancreatic cancer , pancreatic ductal carcinoma , pancreatitis , Perennial Allergic Rhinitis , pituitary-dependent Cushing's disease , polycystic kidney disease , pre-eclampsia , primary pulmonary hypertension , proliferative diabetic retinopathy , psoriasis , Reperfusion Injury , steatotic liver disease , Stomach Neoplasms , Subarachnoid Hemorrhage , systemic lupus erythematosus , T-cell non-Hodgkin lymphoma , Thyroid Neoplasms , transient cerebral ischemia , type 2 diabetes mellitus , urinary bladder cancer Pparg abdominal obesity-metabolic syndrome , acute kidney failure , Acute Lung Injury , acute promyelocytic leukemia , adenocarcinoma , alcoholic cardiomyopathy , Alveolar Bone Loss , Alzheimer's disease , antiphospholipid syndrome , arteriosclerosis , atherosclerosis , autosomal dominant polycystic kidney disease , Barrett's esophagus , cardiovascular system disease , carotid artery disease , Carotid Artery Injuries , Carotid Intimal Medial Thickness 1 , Chemical and Drug Induced Liver Injury , Chronic Allograft Nephropathy , Chronobiology Disorders , cognitive disorder , colon cancer , colon carcinoma , Colonic Neoplasms , Colorectal Neoplasms , congenital generalized lipodystrophy type 2 , congestive heart failure , corneal neovascularization , coronary artery disease , COVID-19 , Crohn's disease , diabetes mellitus , diabetic encephalopathy , Diabetic Nephropathies , diabetic retinopathy , disease of metabolism , Dyslipidemias , end stage renal disease , Endotoxemia , Experimental Autoimmune Myocarditis , Experimental Diabetes Mellitus , Experimental Liver Cirrhosis , Eye Burns , Eye Injuries , familial partial lipodystrophy , familial partial lipodystrophy type 3 , Fetal Growth Retardation , Fibrosis , Follicular Thyroid Cancer , genetic disease , glioblastoma , glomerulonephritis , glomerulosclerosis , Graves ophthalmopathy , hepatocellular carcinoma , high grade glioma , hyperandrogenism , hypertension , Inflammation , Insulin Resistance , ischemia , kidney failure , Left Ventricular Hypertrophy , leukostasis , lipid storage disease , lipodystrophy , Liver Neoplasms , Lymphatic Metastasis , maturity-onset diabetes of the young type 1 , melanoma , metabolic dysfunction-associated steatotic liver disease , Metabolic Syndrome , morbid obesity , Myocardial Reperfusion Injury , Nerve Degeneration , obesity , osteoarthritis , Oxygen-Induced Retinopathy , Pain , pancreatic cancer , pancreatic ductal carcinoma , pancreatitis , Perennial Allergic Rhinitis , pituitary-dependent Cushing's disease , polycystic kidney disease , pre-eclampsia , primary pulmonary hypertension , proliferative diabetic retinopathy , psoriasis , Reperfusion Injury , steatotic liver disease , Stomach Neoplasms , Subarachnoid Hemorrhage , systemic lupus erythematosus , T-cell non-Hodgkin lymphoma , Thyroid Neoplasms , transient cerebral ischemia , type 2 diabetes mellitus , urinary bladder cancer PRDM14 acute lymphoblastic leukemia , lung non-small cell carcinoma , Testicular Germ Cell Tumor Prdm14 acute lymphoblastic leukemia , lung non-small cell carcinoma , Testicular Germ Cell Tumor Prdm14 acute lymphoblastic leukemia , lung non-small cell carcinoma , Testicular Germ Cell Tumor Sin3a Agenesis of Corpus Callosum , autism spectrum disorder , autistic disorder , chromosome 15q24 deletion syndrome , Craniofacial Abnormalities , Dwarfism , Fetal Growth Retardation , genetic disease , Huntington's disease , hydrocephalus , Hyperalgesia , intellectual disability , microcephaly , Neurodevelopmental Disorders , transient cerebral ischemia SIN3A Agenesis of Corpus Callosum , autism spectrum disorder , autistic disorder , chromosome 15q24 deletion syndrome , Craniofacial Abnormalities , Dwarfism , Fetal Growth Retardation , genetic disease , Huntington's disease , hydrocephalus , Hyperalgesia , intellectual disability , microcephaly , Neurodevelopmental Disorders , transient cerebral ischemia Sin3a Agenesis of Corpus Callosum , autism spectrum disorder , autistic disorder , chromosome 15q24 deletion syndrome , Craniofacial Abnormalities , Dwarfism , Fetal Growth Retardation , genetic disease , Huntington's disease , hydrocephalus , Hyperalgesia , intellectual disability , microcephaly , Neurodevelopmental Disorders , transient cerebral ischemia SPI1 acute myeloid leukemia , agammaglobulinemia , agammaglobulinemia 10 , alcohol use disorder , Brain Hypoxia-Ischemia , myeloid leukemia , T-cell acute lymphoblastic leukemia , Transplant Rejection Spi1 acute myeloid leukemia , agammaglobulinemia , agammaglobulinemia 10 , alcohol use disorder , Brain Hypoxia-Ischemia , myeloid leukemia , T-cell acute lymphoblastic leukemia , Transplant Rejection Spi1 acute myeloid leukemia , agammaglobulinemia , agammaglobulinemia 10 , alcohol use disorder , Brain Hypoxia-Ischemia , myeloid leukemia , T-cell acute lymphoblastic leukemia , Transplant Rejection Tet1 autism spectrum disorder , hepatocellular carcinoma TET1 autism spectrum disorder , hepatocellular carcinoma Tet1 autism spectrum disorder , hepatocellular carcinoma Tet2 acute myeloid leukemia , acute promyelocytic leukemia , breast cancer , chronic myelomonocytic leukemia , colon adenocarcinoma , colon adenoma , colorectal cancer , Colorectal Neoplasms , esophagus squamous cell carcinoma , hepatocellular carcinoma , Immunoblastic Lymphadenopathy , Immunodeficiency 75 , lung non-small cell carcinoma , Monocytosis , multiple myeloma , myelodysplastic syndrome , myeloproliferative neoplasm , nasopharynx carcinoma , Neoplasm Metastasis , Neoplasm Recurrence, Local , oral squamous cell carcinoma , peripheral T-cell lymphoma , Prostatic Neoplasms , Refractory Anemia with Excess of Blasts , renal cell carcinoma , Sezary's disease , stomach cancer Tet2 acute myeloid leukemia , acute promyelocytic leukemia , breast cancer , chronic myelomonocytic leukemia , colon adenocarcinoma , colon adenoma , colorectal cancer , Colorectal Neoplasms , esophagus squamous cell carcinoma , hepatocellular carcinoma , Immunoblastic Lymphadenopathy , Immunodeficiency 75 , lung non-small cell carcinoma , Monocytosis , multiple myeloma , myelodysplastic syndrome , myeloproliferative neoplasm , nasopharynx carcinoma , Neoplasm Metastasis , Neoplasm Recurrence, Local , oral squamous cell carcinoma , peripheral T-cell lymphoma , Prostatic Neoplasms , Refractory Anemia with Excess of Blasts , renal cell carcinoma , Sezary's disease , stomach cancer TET2 acute myeloid leukemia , acute promyelocytic leukemia , breast cancer , chronic myelomonocytic leukemia , colon adenocarcinoma , colon adenoma , colorectal cancer , Colorectal Neoplasms , esophagus squamous cell carcinoma , hepatocellular carcinoma , Immunoblastic Lymphadenopathy , Immunodeficiency 75 , lung non-small cell carcinoma , Monocytosis , multiple myeloma , myelodysplastic syndrome , myeloproliferative neoplasm , nasopharynx carcinoma , Neoplasm Metastasis , Neoplasm Recurrence, Local , oral squamous cell carcinoma , peripheral T-cell lymphoma , Prostatic Neoplasms , Refractory Anemia with Excess of Blasts , renal cell carcinoma , Sezary's disease , stomach cancer Tet3 autism spectrum disorder , Beck-Fahrner Syndrome , Developmental Disease , esophagus squamous cell carcinoma , genetic disease , intellectual disability , multiple myeloma , Neurodevelopmental Disorders , NEURODEVELOPMENTAL-CRANIOFACIAL SYNDROME WITH VARIABLE RENAL AND CARDIAC ABNORMALITIES Tet3 autism spectrum disorder , Beck-Fahrner Syndrome , Developmental Disease , esophagus squamous cell carcinoma , genetic disease , intellectual disability , multiple myeloma , Neurodevelopmental Disorders , NEURODEVELOPMENTAL-CRANIOFACIAL SYNDROME WITH VARIABLE RENAL AND CARDIAC ABNORMALITIES TET3 autism spectrum disorder , Beck-Fahrner Syndrome , Developmental Disease , esophagus squamous cell carcinoma , genetic disease , intellectual disability , multiple myeloma , Neurodevelopmental Disorders , NEURODEVELOPMENTAL-CRANIOFACIAL SYNDROME WITH VARIABLE RENAL AND CARDIAC ABNORMALITIES Uhrf1 adenoid cystic carcinoma , Colonic Neoplasms , hepatocellular carcinoma UHRF1 adenoid cystic carcinoma , Colonic Neoplasms , hepatocellular carcinoma Uhrf1 adenoid cystic carcinoma , Colonic Neoplasms , hepatocellular carcinoma Zbtb4 Experimental Liver Cirrhosis ZBTB4 Experimental Liver Cirrhosis Zbtb4 Experimental Liver Cirrhosis Zfp57 diabetes mellitus , genetic disease , transient neonatal diabetes mellitus , transient neonatal diabetes mellitus 1 Zfp57 diabetes mellitus , genetic disease , transient neonatal diabetes mellitus , transient neonatal diabetes mellitus 1 ZFP57 diabetes mellitus , genetic disease , transient neonatal diabetes mellitus , transient neonatal diabetes mellitus 1
abdominal obesity-metabolic syndrome Pparg , PPARG , Pparg Acute Erythroleukemia KMT2A , Kmt2a , Kmt2a acute kidney failure Pparg , PPARG , Pparg Acute Lung Injury Pparg , PPARG , Pparg acute lymphoblastic leukemia Dnmt3a , Dnmt3a , DNMT3A , KDM2B , Kdm2b , Kdm2b , KMT2A , Kmt2a , Kmt2a , Prdm14 , PRDM14 , Prdm14 acute monocytic leukemia Dnmt3a , Dnmt3a , DNMT3A , KMT2A , Kmt2a , Kmt2a acute myeloid leukemia Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , DNMT3A , Dnmt3a , Dnmt3b , DNMT3B , Dnmt3b , Kmt2a , KMT2A , Kmt2a , Kmt2c , Kmt2c , KMT2C , Spi1 , Spi1 , SPI1 , Tet2 , Tet2 , TET2 acute myelomonocytic leukemia KMT2A , Kmt2a , Kmt2a acute promyelocytic leukemia Dnmt3a , Dnmt3a , DNMT3A , Pparg , PPARG , Pparg , Tet2 , Tet2 , TET2 adenocarcinoma Dnmt1 , DNMT1 , Dnmt1 , KMT2A , Kmt2a , Kmt2a , Kmt2c , Kmt2c , KMT2C , Pparg , PPARG , Pparg adenoid cystic carcinoma Kmt2c , Kmt2c , KMT2C , UHRF1 , Uhrf1 , Uhrf1 adult respiratory distress syndrome Ebf1 , Ebf1 , EBF1 agammaglobulinemia Spi1 , Spi1 , SPI1 agammaglobulinemia 10 Spi1 , Spi1 , SPI1 Agenesis of Corpus Callosum SIN3A , Sin3a , Sin3a alcohol use disorder Spi1 , Spi1 , SPI1 alcoholic cardiomyopathy Pparg , PPARG , Pparg alopecia areata Dnmt1 , DNMT1 , Dnmt1 Alveolar Bone Loss Pparg , PPARG , Pparg Alzheimer's disease Pparg , PPARG , Pparg amyotrophic lateral sclerosis Dnmt3a , Dnmt3a , DNMT3A Angelman syndrome Mecp2 , Mecp2 , MECP2 Animal Disease Models Mecp2 , Mecp2 , MECP2 antiphospholipid syndrome Pparg , PPARG , Pparg anxiety disorder Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , Dnmt3b , DNMT3B , Mecp2 , Mecp2 , MECP2 aortic valve stenosis Ogt , OGT , Ogt arteriosclerosis Pparg , PPARG , Pparg asthma Dnmt1 , DNMT1 , Dnmt1 Ataxia Dnmt1 , DNMT1 , Dnmt1 atherosclerosis Pparg , PPARG , Pparg attention deficit hyperactivity disorder Mecp2 , Mecp2 , MECP2 autism spectrum disorder Dnmt1 , DNMT1 , Dnmt1 , DNMT3A , Dnmt3a , Dnmt3a , DNMT3B , Dnmt3b , Dnmt3b , KMT2A , Kmt2a , Kmt2a , Kmt2c , KMT2C , Kmt2c , Mecp2 , Mecp2 , MECP2 , SIN3A , Sin3a , Sin3a , Tet1 , TET1 , Tet1 , TET3 , Tet3 , Tet3 autistic disorder MBD1 , Mbd1 , Mbd1 , MBD4 , Mbd4 , Mbd4 , Mecp2 , Mecp2 , MECP2 , SIN3A , Sin3a , Sin3a autoimmune disease Dnmt1 , DNMT1 , Dnmt1 autoimmune thrombocytopenic purpura Dnmt3a , Dnmt3a , DNMT3A , Dnmt3b , Dnmt3b , DNMT3B autosomal dominant cerebellar ataxia, deafness and narcolepsy Dnmt1 , DNMT1 , Dnmt1 autosomal dominant polycystic kidney disease Pparg , PPARG , Pparg B-Cell Chronic Lymphocytic Leukemia Dnmt3b , Dnmt3b , DNMT3B Barrett's esophagus Pparg , PPARG , Pparg Beck-Fahrner Syndrome Tet3 , TET3 , Tet3 Beckwith-Wiedemann syndrome Dnmt1 , DNMT1 , Dnmt1 Binge Drinking Dnmt1 , DNMT1 , Dnmt1 , Mecp2 , Mecp2 , MECP2 bone marrow disease Dnmt3a , Dnmt3a , DNMT3A Brain Hypoxia-Ischemia Dnmt3a , Dnmt3a , DNMT3A , Mbd2 , MBD2 , Mbd2 , Spi1 , Spi1 , SPI1 Brain Injuries Dnmt1 , DNMT1 , Dnmt1 breast cancer Tet2 , Tet2 , TET2 Breast Neoplasms Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , Dnmt3a , DNMT3A , Dnmt3b , Dnmt3b , DNMT3B , Ogt , OGT , Ogt bronchiolo-alveolar adenocarcinoma Dnmt1 , DNMT1 , Dnmt1 Carcinogenesis Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , Dnmt3a , DNMT3A carcinoma Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , Dnmt3b , DNMT3B Cardiomegaly Ogt , OGT , Ogt cardiomyopathy Dnmt1 , DNMT1 , Dnmt1 cardiovascular system disease Pparg , PPARG , Pparg carotid artery disease Pparg , PPARG , Pparg Carotid Artery Injuries Pparg , PPARG , Pparg Carotid Intimal Medial Thickness 1 Pparg , PPARG , Pparg cerebellar ataxia Dnmt1 , DNMT1 , Dnmt1 cervical cancer KMT2A , Kmt2a , Kmt2a Charcot-Marie-Tooth disease Dnmt1 , DNMT1 , Dnmt1 Chemical and Drug Induced Liver Injury Pparg , PPARG , Pparg cholangiocarcinoma Kmt2c , Kmt2c , KMT2C choline deficiency disease Dnmt1 , DNMT1 , Dnmt1 , Mbd2 , MBD2 , Mbd2 chromosome 15q24 deletion syndrome SIN3A , Sin3a , Sin3a Chromosome Aberrations KMT2A , Kmt2a , Kmt2a Chronic Allograft Nephropathy Pparg , PPARG , Pparg chronic myeloid leukemia Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , Dnmt3a , DNMT3A , Dnmt3b , Dnmt3b , DNMT3B chronic myelomonocytic leukemia Tet2 , Tet2 , TET2 Chronobiology Disorders Pparg , PPARG , Pparg clear cell renal cell carcinoma Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , Dnmt3a , DNMT3A Cocaine-Related Disorders Mecp2 , Mecp2 , MECP2 cognitive disorder Pparg , PPARG , Pparg colon adenocarcinoma Tet2 , Tet2 , TET2 colon adenoma Tet2 , Tet2 , TET2 colon cancer NANOG , Nanog , Nanog , Pparg , PPARG , Pparg colon carcinoma Pparg , PPARG , Pparg Colonic Neoplasms Dnmt1 , DNMT1 , Dnmt1 , Pparg , PPARG , Pparg , UHRF1 , Uhrf1 , Uhrf1 colorectal adenocarcinoma Dnmt3b , Dnmt3b , DNMT3B colorectal cancer Tet2 , Tet2 , TET2 colorectal carcinoma MBD4 , Mbd4 , Mbd4 Colorectal Neoplasms Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , DNMT3B , Dnmt3b , Kdm2a , Kdm2a , KDM2A , MBD4 , Mbd4 , Mbd4 , Pparg , Pparg , PPARG , Tet2 , Tet2 , TET2 congenital disorder of glycosylation Ogt , OGT , Ogt congenital generalized lipodystrophy type 2 Pparg , PPARG , Pparg congenital heart disease Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , Dnmt3a , DNMT3A , Dnmt3b , Dnmt3b , DNMT3B congenital variant of Rett syndrome Mecp2 , Mecp2 , MECP2 congestive heart failure Dnmt1 , DNMT1 , Dnmt1 , Kdm2a , Kdm2a , KDM2A , Ogt , OGT , Ogt , Pparg , PPARG , Pparg corneal neovascularization Pparg , PPARG , Pparg Cornelia de Lange syndrome 1 KMT2A , Kmt2a , Kmt2a coronary artery disease Pparg , PPARG , Pparg COVID-19 Pparg , PPARG , Pparg Craniofacial Abnormalities Dnmt3a , Dnmt3a , DNMT3A , Dnmt3b , Dnmt3b , DNMT3B , Mecp2 , Mecp2 , MECP2 , SIN3A , Sin3a , Sin3a Crohn's disease Dnmt3a , Dnmt3a , DNMT3A , Pparg , PPARG , Pparg dementia Dnmt1 , DNMT1 , Dnmt1 depressive disorder Dnmt3a , Dnmt3a , DNMT3A Desbuquois dysplasia Dnmt3a , Dnmt3a , DNMT3A , KMT2A , Kmt2a , Kmt2a developmental and epileptic encephalopathy 2 Mecp2 , Mecp2 , MECP2 developmental and epileptic encephalopathy 92 Kmt2c , Kmt2c , KMT2C Developmental Disabilities KDM2B , Kdm2b , Kdm2b , Kmt2c , Kmt2c , KMT2C , Mecp2 , Mecp2 , MECP2 Developmental Disease KMT2A , Kmt2a , Kmt2a , Mecp2 , Mecp2 , MECP2 , Tet3 , TET3 , Tet3 diabetes mellitus Pparg , PPARG , Pparg , Zfp57 , Zfp57 , ZFP57 diabetic encephalopathy Pparg , PPARG , Pparg Diabetic Nephropathies Pparg , PPARG , Pparg diabetic retinopathy Pparg , PPARG , Pparg diffuse large B-cell lymphoma Dnmt3a , Dnmt3a , DNMT3A disease of metabolism Pparg , PPARG , Pparg Dwarfism Dnmt3a , Dnmt3a , DNMT3A , SIN3A , Sin3a , Sin3a Dyslipidemias Pparg , PPARG , Pparg Dysmenorrhea Dnmt3b , Dnmt3b , DNMT3B end stage renal disease Pparg , PPARG , Pparg Endometrial Neoplasms Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , Dnmt3b , DNMT3B Endometrioid Carcinomas Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , Dnmt3b , DNMT3B endometriosis Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , Dnmt3a , DNMT3A , Dnmt3b , Dnmt3b , DNMT3B endometriosis of uterus Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , Dnmt3a , DNMT3A , Dnmt3b , Dnmt3b , DNMT3B Endotoxemia Pparg , PPARG , Pparg epilepsy Mecp2 , Mecp2 , MECP2 esophagus squamous cell carcinoma Dnmt3b , DNMT3B , Dnmt3b , Kmt2c , Kmt2c , KMT2C , MBD4 , Mbd4 , Mbd4 , Tet2 , Tet2 , TET2 , Tet3 , TET3 , Tet3 Experimental Arthritis Dnmt1 , DNMT1 , Dnmt1 Experimental Autoimmune Myocarditis Pparg , PPARG , Pparg Experimental Diabetes Mellitus Ogt , OGT , Ogt , Pparg , PPARG , Pparg Experimental Liver Cirrhosis Pparg , PPARG , Pparg , Zbtb4 , Zbtb4 , ZBTB4 Experimental Mammary Neoplasms Dnmt3b , Dnmt3b , DNMT3B Experimental Seizures MBD1 , Mbd1 , Mbd1 , Mbd2 , MBD2 , Mbd2 , Mecp2 , Mecp2 , MECP2 Eye Burns Pparg , PPARG , Pparg Eye Injuries Pparg , PPARG , Pparg Facial Hypertrichosis Mecp2 , Mecp2 , MECP2 Facies Dnmt3a , Dnmt3a , DNMT3A facioscapulohumeral muscular dystrophy 4 Dnmt3b , Dnmt3b , DNMT3B familial partial lipodystrophy Pparg , PPARG , Pparg familial partial lipodystrophy type 3 Pparg , PPARG , Pparg Familial Prostate Cancer Dnmt3b , Dnmt3b , DNMT3B , Kdm2a , Kdm2a , KDM2A , Mbd2 , MBD2 , Mbd2 Female Infertility Dnmt3b , Dnmt3b , DNMT3B fetal alcohol spectrum disorder Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , Dnmt3b , DNMT3B , Mecp2 , Mecp2 , MECP2 Fetal Growth Retardation Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , DNMT3B , Dnmt3b , Mecp2 , MECP2 , Mecp2 , Pparg , Pparg , PPARG , SIN3A , Sin3a , Sin3a Fibrosis Pparg , PPARG , Pparg focal epilepsy Mecp2 , Mecp2 , MECP2 follicular lymphoma Ebf1 , Ebf1 , EBF1 Follicular Thyroid Cancer Pparg , PPARG , Pparg gastric body carcinoma Dnmt3b , Dnmt3b , DNMT3B gastrointestinal system disease Mecp2 , Mecp2 , MECP2 generalized epilepsy Mecp2 , Mecp2 , MECP2 genetic disease Dnmt1 , DNMT1 , Dnmt1 , DNMT3A , Dnmt3a , Dnmt3a , DNMT3B , Dnmt3b , Dnmt3b , KDM2B , Kdm2b , Kdm2b , KMT2A , Kmt2a , Kmt2a , Kmt2c , KMT2C , Kmt2c , MBD4 , Mbd4 , Mbd4 , Mecp2 , Mecp2 , MECP2 , Ogt , OGT , Ogt , Pparg , Pparg , PPARG , SIN3A , Sin3a , Sin3a , TET3 , Tet3 , Tet3 , ZFP57 , Zfp57 , Zfp57 Germ Cell and Embryonal Neoplasms NANOG , Nanog , Nanog glioblastoma Dnmt3a , Dnmt3a , DNMT3A , Pparg , PPARG , Pparg glomerulonephritis Pparg , PPARG , Pparg glomerulosclerosis Pparg , PPARG , Pparg Graves ophthalmopathy Pparg , PPARG , Pparg Graves' disease Dnmt1 , DNMT1 , Dnmt1 Growth Disorders Dnmt3a , Dnmt3a , DNMT3A head and neck squamous cell carcinoma Dnmt3b , Dnmt3b , DNMT3B Hearing Loss Dnmt1 , DNMT1 , Dnmt1 , Mecp2 , Mecp2 , MECP2 hepatocellular carcinoma Dnmt1 , DNMT1 , Dnmt1 , DNMT3A , Dnmt3a , Dnmt3a , DNMT3B , Dnmt3b , Dnmt3b , KMT2A , Kmt2a , Kmt2a , Kmt2c , KMT2C , Kmt2c , Mecp2 , Mecp2 , MECP2 , Pparg , Pparg , PPARG , Tet1 , TET1 , Tet1 , Tet2 , TET2 , Tet2 , UHRF1 , Uhrf1 , Uhrf1 Hereditary Neoplastic Syndromes MBD4 , Mbd4 , Mbd4 hereditary sensory neuropathy Dnmt1 , DNMT1 , Dnmt1 hereditary sensory neuropathy type 1E Dnmt1 , DNMT1 , Dnmt1 Heyn-Sproul-Jackson Syndrome Dnmt3a , Dnmt3a , DNMT3A high grade glioma Pparg , PPARG , Pparg Hirsutism KMT2A , Kmt2a , Kmt2a Huntington's disease SIN3A , Sin3a , Sin3a hydrocephalus SIN3A , Sin3a , Sin3a Hyperalgesia Mecp2 , Mecp2 , MECP2 , SIN3A , Sin3a , Sin3a hyperandrogenism Pparg , PPARG , Pparg hypertension Pparg , PPARG , Pparg hypertrophic cardiomyopathy Dnmt1 , DNMT1 , Dnmt1 Immunoblastic Lymphadenopathy Dnmt3a , Dnmt3a , DNMT3A , Tet2 , Tet2 , TET2 Immunodeficiency 75 Tet2 , Tet2 , TET2 immunodeficiency-centromeric instability-facial anomalies syndrome Dnmt3b , Dnmt3b , DNMT3B immunodeficiency-centromeric instability-facial anomalies syndrome 1 Dnmt3b , Dnmt3b , DNMT3B Inflammation Pparg , PPARG , Pparg Insulin Resistance Pparg , PPARG , Pparg intellectual disability Dnmt3a , DNMT3A , Dnmt3a , Kmt2a , KMT2A , Kmt2a , Kmt2c , Kmt2c , KMT2C , Mecp2 , MECP2 , Mecp2 , SIN3A , Sin3a , Sin3a , Tet3 , TET3 , Tet3 Intestinal Neoplasms MBD4 , Mbd4 , Mbd4 ischemia Pparg , PPARG , Pparg juvenile myelomonocytic leukemia Dnmt3a , Dnmt3a , DNMT3A Kabuki Syndrome 1 Dnmt3b , Dnmt3b , DNMT3B , KMT2A , Kmt2a , Kmt2a kidney failure Pparg , PPARG , Pparg Kidney Reperfusion Injury KMT2A , Kmt2a , Kmt2a Klatskin's tumor Kmt2c , Kmt2c , KMT2C Kleefstra syndrome 1 Kmt2c , Kmt2c , KMT2C Kleefstra syndrome 2 Kmt2c , Kmt2c , KMT2C language disorder KMT2A , Kmt2a , Kmt2a laryngeal carcinoma Kmt2c , Kmt2c , KMT2C learning disability Mecp2 , Mecp2 , MECP2 Left Ventricular Hypertrophy Pparg , PPARG , Pparg leukemia KMT2A , Kmt2a , Kmt2a leukostasis Pparg , PPARG , Pparg lipid storage disease Pparg , PPARG , Pparg lipodystrophy Ebf1 , Ebf1 , EBF1 , Pparg , PPARG , Pparg liver cirrhosis Dnmt1 , DNMT1 , Dnmt1 Liver Neoplasms Pparg , PPARG , Pparg lung adenocarcinoma Dnmt3a , Dnmt3a , Dnmt3b , Dnmt3b , DNMT3B , Kmt2c , Kmt2c , KMT2C lung cancer Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , DNMT3A , Dnmt3a , Dnmt3b , DNMT3B , Dnmt3b , MBD1 , Mbd1 , Mbd1 , MBD4 , Mbd4 , Mbd4 lung carcinoma Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , Dnmt3a , DNMT3A Lung Neoplasms Dnmt3a , Dnmt3a , DNMT3A lung non-small cell carcinoma Dnmt1 , DNMT1 , Dnmt1 , Kdm2a , Kdm2a , KDM2A , Kmt2c , Kmt2c , KMT2C , Prdm14 , PRDM14 , Prdm14 , Tet2 , Tet2 , TET2 lung small cell carcinoma Dnmt3b , Dnmt3b , DNMT3B , KMT2A , Kmt2a , Kmt2a Lymphatic Metastasis Dnmt3b , Dnmt3b , DNMT3B , Pparg , PPARG , Pparg lymphoid leukemia KMT2A , Kmt2a , Kmt2a macular degeneration Mbd2 , MBD2 , Mbd2 maturity-onset diabetes of the young type 1 Pparg , PPARG , Pparg medulloblastoma Dnmt1 , DNMT1 , Dnmt1 melanoma Dnmt3a , Dnmt3a , DNMT3A , Pparg , PPARG , Pparg metabolic dysfunction and alcohol associated liver disease Dnmt1 , DNMT1 , Dnmt1 metabolic dysfunction-associated steatotic liver disease Pparg , PPARG , Pparg Metabolic Syndrome Pparg , PPARG , Pparg microcephaly Dnmt3a , DNMT3A , Dnmt3a , Kmt2a , KMT2A , Kmt2a , Kmt2c , Kmt2c , KMT2C , Mecp2 , MECP2 , Mecp2 , SIN3A , Sin3a , Sin3a Micrognathism Mecp2 , Mecp2 , MECP2 middle cerebral artery infarction Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , Dnmt3a , DNMT3A Monocytosis Tet2 , Tet2 , TET2 morbid obesity Pparg , PPARG , Pparg mucoepidermoid carcinoma Dnmt1 , DNMT1 , Dnmt1 multiple myeloma Dnmt3a , Dnmt3a , DNMT3A , Kmt2c , Kmt2c , KMT2C , Tet2 , Tet2 , TET2 , Tet3 , TET3 , Tet3 Multiple Primary Neoplasms Dnmt3b , Dnmt3b , DNMT3B Muscle Hypotonia Dnmt3a , Dnmt3a , DNMT3A , Mecp2 , Mecp2 , MECP2 myelodysplastic syndrome Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , Dnmt3a , DNMT3A , Dnmt3b , Dnmt3b , DNMT3B , Tet2 , Tet2 , TET2 myelofibrosis KMT2A , Kmt2a , Kmt2a myeloid leukemia KMT2A , Kmt2a , Kmt2a , Spi1 , Spi1 , SPI1 myeloid neoplasm Dnmt3a , Dnmt3a , DNMT3A myeloproliferative neoplasm Tet2 , Tet2 , TET2 Myocardial Reperfusion Injury Pparg , PPARG , Pparg Myoclonus Mecp2 , Mecp2 , MECP2 nasopharynx carcinoma Kmt2c , Kmt2c , KMT2C , Tet2 , Tet2 , TET2 Neoplasm Invasiveness Dnmt1 , DNMT1 , Dnmt1 Neoplasm Metastasis Dnmt3b , Dnmt3b , DNMT3B , Tet2 , Tet2 , TET2 Neoplasm Recurrence, Local Dnmt3a , Dnmt3a , DNMT3A , Tet2 , Tet2 , TET2 Neoplastic Cell Transformation Ebf1 , Ebf1 , EBF1 , NANOG , Nanog , Nanog Nerve Degeneration Pparg , PPARG , Pparg Nervous System Malformations Dnmt3a , Dnmt3a , DNMT3A , Mecp2 , Mecp2 , MECP2 neural tube defect KDM2B , Kdm2b , Kdm2b Neurodevelopmental Disorders Dnmt3a , DNMT3A , Dnmt3a , Kdm2a , Kdm2a , KDM2A , KDM2B , Kdm2b , Kdm2b , Kmt2a , KMT2A , Kmt2a , Kmt2c , Kmt2c , KMT2C , Mecp2 , MECP2 , Mecp2 , SIN3A , Sin3a , Sin3a , Tet3 , TET3 , Tet3 NEURODEVELOPMENTAL-CRANIOFACIAL SYNDROME WITH VARIABLE RENAL AND CARDIAC ABNORMALITIES Tet3 , TET3 , Tet3 non-Hodgkin lymphoma KMT2A , Kmt2a , Kmt2a non-syndromic X-linked intellectual disability Mecp2 , Mecp2 , MECP2 non-syndromic X-linked intellectual disability 106 Ogt , OGT , Ogt obesity Dnmt3a , Dnmt3a , DNMT3A , Pparg , PPARG , Pparg Oral Lichen Planus Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , Dnmt3b , DNMT3B oral squamous cell carcinoma Dnmt3b , Dnmt3b , DNMT3B , Tet2 , Tet2 , TET2 osteoarthritis Pparg , PPARG , Pparg Ovarian Neoplasms Dnmt1 , DNMT1 , Dnmt1 Oxygen-Induced Retinopathy Pparg , PPARG , Pparg Pain Pparg , PPARG , Pparg pancreatic cancer Pparg , PPARG , Pparg pancreatic ductal carcinoma Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , Dnmt3b , DNMT3B , KDM2B , Kdm2b , Kdm2b , Pparg , PPARG , Pparg Pancreatic Intraepithelial Neoplasia Dnmt1 , DNMT1 , Dnmt1 pancreatitis Pparg , PPARG , Pparg Parasitic Liver Diseases Kmt2c , Kmt2c , KMT2C Parkinson's disease NANOG , Nanog , Nanog Perennial Allergic Rhinitis Pparg , PPARG , Pparg peripheral T-cell lymphoma Dnmt3a , Dnmt3a , DNMT3A , Tet2 , Tet2 , TET2 Peritoneal Fibrosis Dnmt1 , DNMT1 , Dnmt1 periventricular nodular heterotopia Mecp2 , Mecp2 , MECP2 pervasive developmental disorder Mecp2 , Mecp2 , MECP2 Pituitary Stalk Interruption Syndrome Dnmt1 , DNMT1 , Dnmt1 pituitary-dependent Cushing's disease Pparg , PPARG , Pparg polycystic kidney disease Pparg , PPARG , Pparg pre-eclampsia Pparg , PPARG , Pparg primary cutaneous T-cell non-Hodgkin lymphoma Dnmt3a , Dnmt3a , DNMT3A primary immunodeficiency disease Dnmt3b , Dnmt3b , DNMT3B primary pulmonary hypertension Pparg , PPARG , Pparg proliferative diabetic retinopathy Pparg , PPARG , Pparg prostate adenocarcinoma Dnmt1 , DNMT1 , Dnmt1 , Dnmt3a , Dnmt3a , DNMT3A , Dnmt3b , Dnmt3b , DNMT3B prostate cancer Dnmt1 , DNMT1 , Dnmt1 , KMT2A , Kmt2a , Kmt2a , Kmt2c , Kmt2c , KMT2C prostate carcinoma in situ Dnmt1 , DNMT1 , Dnmt1 Prostatic Neoplasms Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , DNMT3B , Dnmt3b , Kmt2a , KMT2A , Kmt2a , Kmt2c , Kmt2c , KMT2C , MBD1 , Mbd1 , Mbd1 , Mbd2 , MBD2 , Mbd2 , Tet2 , Tet2 , TET2 prostatitis Dnmt3a , Dnmt3a , DNMT3A , Dnmt3b , Dnmt3b , DNMT3B psoriasis Pparg , PPARG , Pparg Psychomotor Disorders Mecp2 , Mecp2 , MECP2 pulmonary fibrosis Mecp2 , Mecp2 , MECP2 Refractory Anemia with Excess of Blasts Tet2 , Tet2 , TET2 renal cell carcinoma Tet2 , Tet2 , TET2 Reperfusion Injury Pparg , PPARG , Pparg respiratory failure Mecp2 , Mecp2 , MECP2 Rett syndrome Mecp2 , Mecp2 , MECP2 Rett Syndrome, Atypical Mecp2 , Mecp2 , MECP2 Rett Syndrome, Zappella Variant Mecp2 , Mecp2 , MECP2 rheumatoid arthritis Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , DNMT3B , Dnmt3b , Kmt2c , Kmt2c , KMT2C , Mbd2 , MBD2 , Mbd2 , MBD4 , Mbd4 , Mbd4 Rubinstein Taybi like Syndrome KMT2A , Kmt2a , Kmt2a schizophrenia Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , DNMT3B , Dnmt3b , KDM2B , Kdm2b , Kdm2b , Mbd2 , MBD2 , Mbd2 , Mecp2 , MECP2 , Mecp2 sciatic neuropathy Dnmt3a , Dnmt3a , DNMT3A severe combined immunodeficiency KDM2B , Kdm2b , Kdm2b severe congenital encephalopathy due to MECP2 mutation Mecp2 , Mecp2 , MECP2 Sezary's disease Dnmt3a , Dnmt3a , DNMT3A , Kmt2c , Kmt2c , KMT2C , Tet2 , Tet2 , TET2 skin melanoma Dnmt3b , Dnmt3b , DNMT3B Smith-Magenis syndrome Mecp2 , Mecp2 , MECP2 spastic ataxia Dnmt1 , DNMT1 , Dnmt1 Spastic Paraparesis Dnmt1 , DNMT1 , Dnmt1 squamous cell carcinoma Kmt2c , Kmt2c , KMT2C steatotic liver disease Pparg , PPARG , Pparg stereotypic movement disorder Mecp2 , Mecp2 , MECP2 stomach cancer Dnmt3a , Dnmt3a , DNMT3A , Kmt2c , Kmt2c , KMT2C , Tet2 , Tet2 , TET2 stomach carcinoma Kmt2c , Kmt2c , KMT2C Stomach Neoplasms Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , DNMT3B , Dnmt3b , Kmt2a , KMT2A , Kmt2a , Kmt2c , Kmt2c , KMT2C , Pparg , Pparg , PPARG Subarachnoid Hemorrhage Pparg , PPARG , Pparg syndromic intellectual disability Kmt2c , Kmt2c , KMT2C syndromic X-linked intellectual disability Lubs type Mecp2 , Mecp2 , MECP2 systemic lupus erythematosus Dnmt1 , DNMT1 , Dnmt1 , Pparg , PPARG , Pparg T-cell acute lymphoblastic leukemia Dnmt3a , Dnmt3a , DNMT3A , Spi1 , Spi1 , SPI1 T-cell non-Hodgkin lymphoma KDM2B , Kdm2b , Kdm2b , Pparg , PPARG , Pparg Tatton-Brown-Rahman syndrome Dnmt3a , Dnmt3a , DNMT3A teratoma NANOG , Nanog , Nanog Testicular Germ Cell Tumor Prdm14 , PRDM14 , Prdm14 tetralogy of Fallot KMT2A , Kmt2a , Kmt2a thymoma Dnmt3b , Dnmt3b , DNMT3B Thyroid Neoplasms Pparg , PPARG , Pparg transient cerebral ischemia Kmt2a , KMT2A , Kmt2a , MBD1 , Mbd1 , Mbd1 , Mbd2 , MBD2 , Mbd2 , Mecp2 , MECP2 , Mecp2 , Pparg , Pparg , PPARG , SIN3A , Sin3a , Sin3a transient neonatal diabetes mellitus Zfp57 , Zfp57 , ZFP57 transient neonatal diabetes mellitus 1 Zfp57 , Zfp57 , ZFP57 transitional cell carcinoma KMT2A , Kmt2a , Kmt2a , Kmt2c , Kmt2c , KMT2C Transplant Rejection Spi1 , Spi1 , SPI1 Tumor Predisposition Syndrome 2 MBD4 , Mbd4 , Mbd4 type 1 diabetes mellitus Dnmt3b , Dnmt3b , DNMT3B , Mbd2 , MBD2 , Mbd2 type 2 diabetes mellitus Dnmt1 , DNMT1 , Dnmt1 , Pparg , PPARG , Pparg urinary bladder cancer Dnmt1 , DNMT1 , Dnmt1 , KMT2A , Kmt2a , Kmt2a , Kmt2c , Kmt2c , KMT2C , Pparg , PPARG , Pparg Uterine Cervical Neoplasms Dnmt1 , DNMT1 , Dnmt1 , Dnmt3b , Dnmt3b , DNMT3B uveal melanoma MBD4 , Mbd4 , Mbd4 Wiedemann-Steiner syndrome KMT2A , Kmt2a , Kmt2a X-Linked Intellectual Developmental Disorders Mecp2 , Mecp2 , MECP2 X-linked intellectual disability-psychosis-macroorchidism syndrome Mecp2 , Mecp2 , MECP2