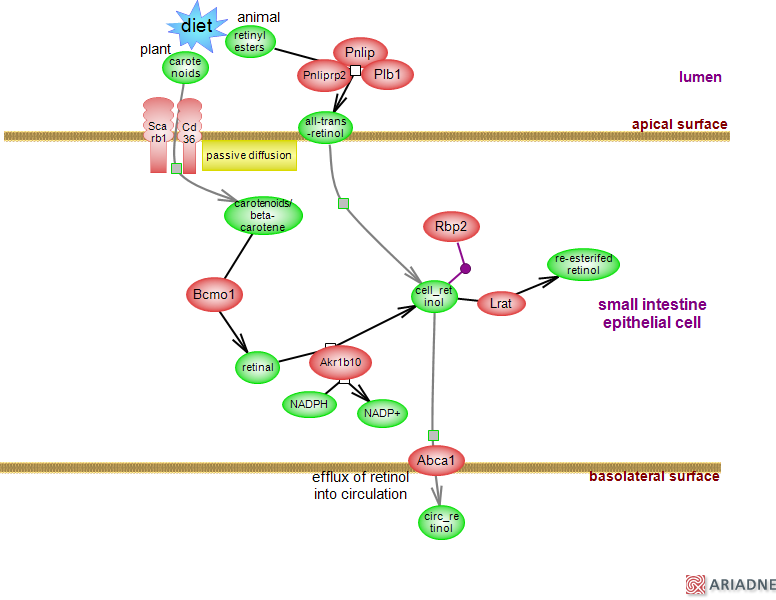
Genes in Pathway:
|
|
| G
|
Abca1
|
ATP binding cassette subfamily A member 1
|
|
ISO
|
|
RGD |
PMID:21718801 |
RGD:6484679 |
NCBI chr 5:72,473,676...72,596,563
Ensembl chr 5:72,473,676...72,596,473
|

|
| G
|
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 2:229,622,092...229,640,120
Ensembl chr 2:229,622,095...229,641,879
|

|
| G
|
Adh5
|
alcohol dehydrogenase 5 (class III), chi polypeptide
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 2:229,648,557...229,660,964
Ensembl chr 2:229,648,501...229,665,232
|

|
| G
|
Adh6
|
alcohol dehydrogenase 6 (class V)
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 2:229,576,612...229,608,359
Ensembl chr 2:229,576,636...229,607,914
|

|
| G
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 2:229,422,125...229,436,584
Ensembl chr 2:229,421,252...229,436,691
|

|
| G
|
Akr1b10
|
aldo-keto reductase family 1 member B10
|
|
IDA
ISO
|
|
RGD |
PMID:20709016 PMID:16970545 |
RGD:6903952, RGD:6903957 |
NCBI chr 4:64,005,632...64,027,860
Ensembl chr 4:64,007,420...64,020,847
|

|
| G
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr 1:227,426,939...227,579,497
Ensembl chr 1:227,427,070...227,579,500
|

|
| G
|
Aldh1a2
|
aldehyde dehydrogenase 1 family, member A2
|
|
IDA
IEA
ISO
|
|
KEGG
SMPDB
RGD |
PMID:12547725 |
SMP:00074 rno:00830, RGD:1576367 |
NCBI chr 8:80,758,641...80,837,891
Ensembl chr 8:80,758,142...80,837,883
|

|
| G
|
Awat1
|
acyl-CoA wax alcohol acyltransferase 1
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr X:69,708,286...69,714,531
Ensembl chr X:69,708,286...69,714,531
|

|
| G
|
Awat2
|
acyl-CoA wax alcohol acyltransferase 2
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr X:69,521,778...69,530,664
Ensembl chr X:69,521,778...69,530,664
|

|
| G
|
Bco1
|
beta-carotene oxygenase 1
|
|
ISO
TAS
IEA
|
|
KEGG
SMPDB
RGD |
PMID:21569862 PMID:21718801 |
SMP:00074 rno:00830, RGD:6903956, RGD:6484679 |
NCBI chr19:62,058,061...62,094,923
Ensembl chr19:62,058,089...62,094,917
|

|
| G
|
Cd36
|
CD36 molecule
|
|
ISO
|
|
RGD |
PMID:21718801 |
RGD:6484679 |
NCBI chr 4:18,209,088...18,302,142
|

|
| G
|
Cyp1a1
|
cytochrome P450, family 1, subfamily a, polypeptide 1
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 8:66,991,940...66,998,014
Ensembl chr 8:66,991,970...66,998,012
|

|
| G
|
Cyp1a2
|
cytochrome P450, family 1, subfamily a, polypeptide 2
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 8:66,971,261...66,978,149
Ensembl chr 8:66,971,261...66,978,149
|

|
| G
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 1:244,883,822...244,887,657
Ensembl chr 1:244,883,752...244,887,657
|

|
| G
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 4:118,599,356...118,616,176
Ensembl chr 4:118,599,356...118,616,176
|

|
| G
|
Cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:244,870,412...244,881,610
Ensembl chr 1:244,871,612...244,880,297
|

|
| G
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:91,359,278...91,372,554
Ensembl chr 1:91,359,272...91,389,730
|

|
| G
|
Cyp2a2
|
cytochrome P450, family 2, subfamily a, polypeptide 2
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:91,391,492...91,417,158
Ensembl chr 1:91,391,502...91,417,158
|

|
| G
|
Cyp2a3
|
cytochrome P450, family 2, subfamily a, polypeptide 3
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 1:91,299,584...91,307,650
Ensembl chr 1:91,299,561...91,307,649
|

|
| G
|
Cyp2b1
|
cytochrome P450, family 2, subfamily b, polypeptide 1
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:90,646,098...90,669,762
|

|
| G
|
Cyp2b12
|
cytochrome P450, family 2, subfamily b, polypeptide 12
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:91,135,294...91,147,094
Ensembl chr 1:91,135,294...91,208,159
|

|
| G
|
Cyp2b2
|
cytochrome P450, family 2, subfamily b, polypeptide 2
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:90,722,243...90,736,272
|

|
| G
|
Cyp2b21
|
cytochrome P450, family 2, subfamily b, polypeptide 21
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:91,013,781...91,042,752
Ensembl chr 1:91,013,782...91,042,752
|

|
| G
|
Cyp2b3
|
cytochrome P450, family 2, subfamily b, polypeptide 3
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 1:90,780,468...90,859,852
|

|
| G
|
Cyp2c11
|
cytochrome P450, subfamily 2, polypeptide 11
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:246,175,216...246,211,445
Ensembl chr 1:246,174,711...246,211,853
|

|
| G
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:248,619,887...248,681,945
Ensembl chr 1:248,619,882...248,677,042
|

|
| G
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:248,706,787...248,787,239
Ensembl chr 1:248,706,788...248,787,210
|

|
| G
|
Cyp2c22
|
cytochrome P450, family 2, subfamily c, polypeptide 22
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:248,941,143...248,980,767
Ensembl chr 1:248,904,062...248,971,182
|

|
| G
|
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:252,838,427...252,863,081
Ensembl chr 1:252,838,433...252,863,067
|

|
| G
|
Cyp2c7
|
cytochrome P450, family 2, subfamily c, polypeptide 7
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 1:247,966,295...248,022,393
Ensembl chr 1:247,966,346...248,022,388
|

|
| G
|
Cyp3a18
|
cytochrome P450, family 3, subfamily a, polypeptide 18
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr12:13,994,312...14,044,185
Ensembl chr12:13,994,311...14,045,788
|

|
| G
|
Cyp3a2
|
cytochrome P450, family 3, subfamily a, polypeptide 2
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr12:14,321,771...14,343,886
Ensembl chr12:14,321,771...14,343,857
|

|
| G
|
Cyp3a23-3a1
|
cytochrome P450, family 3, subfamily a, polypeptide 23-polypeptide 1
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr12:14,369,950...14,398,803
Ensembl chr12:14,368,266...14,398,813
|

|
| G
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr12:21,919,955...21,960,160
Ensembl chr12:21,919,940...21,960,154
|

|
| G
|
Cyp4a1
|
cytochrome P450, family 4, subfamily a, polypeptide 1
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 5:134,360,096...134,374,233
Ensembl chr 5:134,360,111...134,374,231
|

|
| G
|
Cyp4a2
|
cytochrome P450, family 4, subfamily a, polypeptide 2
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 5:134,160,356...134,170,908
Ensembl chr 5:134,160,358...134,176,156
|

|
| G
|
Cyp4a3
|
cytochrome P450, family 4, subfamily a, polypeptide 3
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 5:134,334,396...134,352,268
Ensembl chr 5:134,334,701...134,352,236
|

|
| G
|
Cyp4a8
|
cytochrome P450, family 4, subfamily a, polypeptide 8
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 5:133,938,882...133,970,226
Ensembl chr 5:133,938,883...133,970,226
|

|
| G
|
Dgat1
|
diacylglycerol O-acyltransferase 1
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 7:110,104,514...110,119,091
Ensembl chr 7:110,098,906...110,115,016
|

|
| G
|
Dhrs3
|
dehydrogenase/reductase 3
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 5:162,031,853...162,065,655
Ensembl chr 5:162,031,386...162,065,653
|

|
| G
|
Dhrs4
|
dehydrogenase/reductase 4
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr15:32,924,482...32,948,103
Ensembl chr15:32,936,522...32,948,101
|

|
| G
|
Dhrs9
|
dehydrogenase/reductase 9
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 3:74,553,357...74,579,535
Ensembl chr 3:74,555,606...74,581,376
|

|
| G
|
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr11:91,656,334...91,672,800
Ensembl chr11:91,656,335...91,672,800
|

|
| G
|
Erp29
|
endoplasmic reticulum protein 29
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr12:40,750,177...40,756,607
Ensembl chr12:40,750,314...40,757,010
|

|
| G
|
Hsp90b1
|
heat shock protein 90 beta family member 1
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr 7:22,997,962...23,012,293
Ensembl chr 7:22,997,962...23,012,293
|

|
| G
|
Hspa5
|
heat shock protein family A (Hsp70) member 5
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr 3:38,453,016...38,457,475
Ensembl chr 3:38,453,041...38,458,790
|

|
| G
|
Hyou1
|
hypoxia up-regulated 1
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr 8:53,602,882...53,614,998
Ensembl chr 8:53,602,604...53,614,997
|

|
| G
|
Lrat
|
lecithin retinol acyltransferase
|
|
TAS
IEA
ISO
|
|
KEGG
SMPDB
RGD |
PMID:21718801 |
SMP:00074 rno:00830, RGD:6484679 |
NCBI chr 2:170,562,148...170,571,212
Ensembl chr 2:170,565,543...170,571,148
|

|
| G
|
Pdia2
|
protein disulfide isomerase family A, member 2
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr10:15,720,301...15,723,386
Ensembl chr10:15,720,301...15,723,396
|

|
| G
|
Pdia4
|
protein disulfide isomerase family A, member 4
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr 4:78,134,144...78,153,191
Ensembl chr 4:78,134,144...78,153,191
|

|
| G
|
Pdia6
|
protein disulfide isomerase family A, member 6
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr 6:45,791,704...45,808,947
Ensembl chr 6:45,791,678...45,810,091
|

|
| G
|
Plb1
|
phospholipase B1
|
|
IDA
TAS
|
|
RGD |
PMID:8117729 PMID:21718801 |
RGD:6903951, RGD:6484679 |
NCBI chr 6:29,807,127...29,945,607
Ensembl chr 6:29,809,290...29,930,183
|

|
| G
|
Pnlip
|
pancreatic lipase
|
|
TAS
|
|
RGD |
PMID:21718801 |
RGD:6484679 |
NCBI chr 1:267,760,073...267,797,797
Ensembl chr 1:267,784,724...267,797,795
|

|
| G
|
Pnliprp2
|
pancreatic lipase related protein 2
|
|
ISO
|
|
RGD |
PMID:21718801 |
RGD:6484679 |
NCBI chr 1:267,883,906...267,902,572
Ensembl chr 1:267,883,906...267,902,572
|

|
| G
|
Pnpla4
|
patatin like domain 4, phospholipase and triacylglycerol lipase
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr X:46,200,351...46,205,517
Ensembl chr X:46,193,332...46,205,089
|

|
| G
|
Ppib
|
peptidylprolyl isomerase B
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr 8:75,498,966...75,504,823
Ensembl chr 8:75,498,797...75,504,847
|

|
| G
|
Rbp2
|
retinol binding protein 2
|
|
ISO
|
|
RGD |
PMID:21718801 |
RGD:6484679 |
NCBI chr 8:107,958,670...107,983,850
|

|
| G
|
Rdh10
|
retinol dehydrogenase 10
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 5:7,969,931...7,998,834
Ensembl chr 5:7,971,705...7,999,019
|

|
| G
|
Rdh11
|
retinol dehydrogenase 11
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 6:103,712,373...103,728,247
Ensembl chr 6:103,712,374...103,729,741
|

|
| G
|
Rdh12
|
retinol dehydrogenase 12
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 6:103,748,457...103,761,379
Ensembl chr 6:103,748,427...103,761,380
|

|
| G
|
Rdh16
|
retinol dehydrogenase 16
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr 7:65,499,763...65,510,304
|

|
| G
|
Rdh5
|
retinol dehydrogenase 5
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 7:1,912,120...1,931,836
Ensembl chr 7:1,925,335...1,936,049
|

|
| G
|
Rdh7
|
retinol dehydrogenase 7
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 7:65,561,259...65,574,618
Ensembl chr 7:65,560,822...65,574,635
|

|
| G
|
Rdh8
|
retinol dehydrogenase 8
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 8:27,629,380...27,636,651
Ensembl chr 8:27,629,380...27,636,651
|

|
| G
|
Retsat
|
retinol saturase
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 4:106,211,459...106,220,222
Ensembl chr 4:106,211,436...106,229,552
|

|
| G
|
Rpe65
|
retinoid isomerohydrolase RPE65
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 2:251,425,228...251,457,209
Ensembl chr 2:251,425,368...251,457,156
|

|
| G
|
Scarb1
|
scavenger receptor class B, member 1
|
|
ISO
|
|
RGD |
PMID:21718801 |
RGD:6484679 |
NCBI chr12:36,957,302...37,023,982
Ensembl chr12:36,957,496...37,023,980
|

|
| G
|
Sdf2l1
|
stromal cell-derived factor 2-like 1
|
|
ISO
|
|
SMPDB |
|
SMP:00074 |
NCBI chr11:97,376,878...97,379,120
Ensembl chr11:97,376,878...97,379,120
|

|
| G
|
Ugt1a1
|
UDP glucuronosyltransferase family 1 member A1
|
|
IEA
ISO
|
|
KEGG
SMPDB |
|
SMP:00074 rno:00830 |
NCBI chr 9:96,249,143...96,256,264
Ensembl chr 9:96,144,786...96,256,264
|

|
| G
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 9:96,239,019...96,256,264
Ensembl chr 9:96,144,786...96,256,264
|

|
| G
|
Ugt1a3
|
UDP glycosyltransferase 1 family, polypeptide A3
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 9:96,228,134...96,256,264
Ensembl chr 9:96,144,786...96,256,264
|

|
| G
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 9:96,210,053...96,256,264
Ensembl chr 9:96,144,786...96,256,264
|

|
| G
|
Ugt1a6a
|
UDP glucuronosyltransferase family 1 member A6a
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 9:96,195,018...96,256,264
Ensembl chr 9:96,144,786...96,256,264
|

|
| G
|
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 9:96,187,382...96,256,264
Ensembl chr 9:96,144,786...96,256,264
|

|
| G
|
Ugt1a8
|
UDP glucuronosyltransferase family 1 member A8
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 9:96,174,899...96,256,264
Ensembl chr 9:96,144,786...96,256,264
|

|
| G
|
Ugt1a9
|
UDP glucuronosyltransferase family 1 member A9
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr 9:96,144,786...96,256,264
Ensembl chr 9:96,144,786...96,256,264
|

|
| G
|
Ugt2a1
|
UDP glucuronosyltransferase family 2 member A1
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr14:20,797,262...20,825,111
Ensembl chr14:20,800,263...20,825,111
|

|
| G
|
Ugt2a3
|
UDP glucuronosyltransferase family 2 member A3
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr14:20,851,970...20,869,974
Ensembl chr14:20,851,969...20,869,973
|

|
| G
|
Ugt2b
|
UDP glycosyltransferase 2 family, polypeptide B
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr14:20,985,449...21,006,267
Ensembl chr14:20,985,108...21,020,850
|

|
| G
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr14:21,378,882...21,390,631
Ensembl chr14:21,378,853...21,392,331
|

|
| G
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37
|
|
IEA
|
|
KEGG |
|
rno:00830 |
NCBI chr14:21,172,136...21,188,768
Ensembl chr14:21,172,133...21,190,661
|

|
|