NICOTINAMIDE ADENINE DINUCLEOTIDE METABOLIC PATHWAY (PW:0002580)
Description
Nicotinamide adenine dinucleotide (NAD+) is an essential coenzyme of enzyme-catalyzed reduction-oxidation (redox) reactions. Its discovery and mechanistic understanding span more than a century and brought four Nobel prizes. NAD+ and its reduced form NADH, and the phosphorylated NADP/NADPH forms, serve as hydride acceptors or donors in over 2,000 redox reactions. In addition to its known role as a cofactor in redox reactions, NAD+ serves as a substrate for several classes of enzymes that include the sirtuin deacetylases, the adenosine diphosphate (ADP)-ribose transferases/poly(ADP-ribose) polymerases, and the cyclic ADP-ribose (cADPR) synthases. The ensuing protein modification and messenger generation play a spectrum of roles in gene-regulatory, signaling, metabolic and cellular homeostasis pathways, and also in aging and diseases. Unlike the reversible shuttling between oxidized and reduced forms, which do not change the overall coenzyme concentration, the activity of NAD+ consuming enzymes requires the replenishing of the NAD+ pool. This heightens the importance of NAD+ biosynthesis and of its regulation. Several routes exist to synthesize NAD+ from various dietary sources. A
de novo pathway, downstream of kynurenine metabolism of tryptophan degradation, uses quinolinic acid (QUIN) as the starting material. Others, described as salvage pathways and the primary source of NAD+, use dietary niacin (vitamin B3) - nicotinic acid (NA) and its pyridine-nucleoside or amide forms of nicotinamide riboside (NR) and nicotinamide (NAM), respectively. NAM is also the product of the reactions carried out by the enzymes that use NAD+ as a substrate and which split the molecule at its N-glycosidic bond. The uptake of NAD precursors into the cell is not yet well characterized but several transporters for tryptophan and nicotinic acid have been reported. Transport of NAD and/or metabolites across the plasma membrane is also not well understood. Connexin 43 (Gja1) hemichannels may mediate flux of NAD while the purinergic P2x7 receptor may mediate NADH transport.
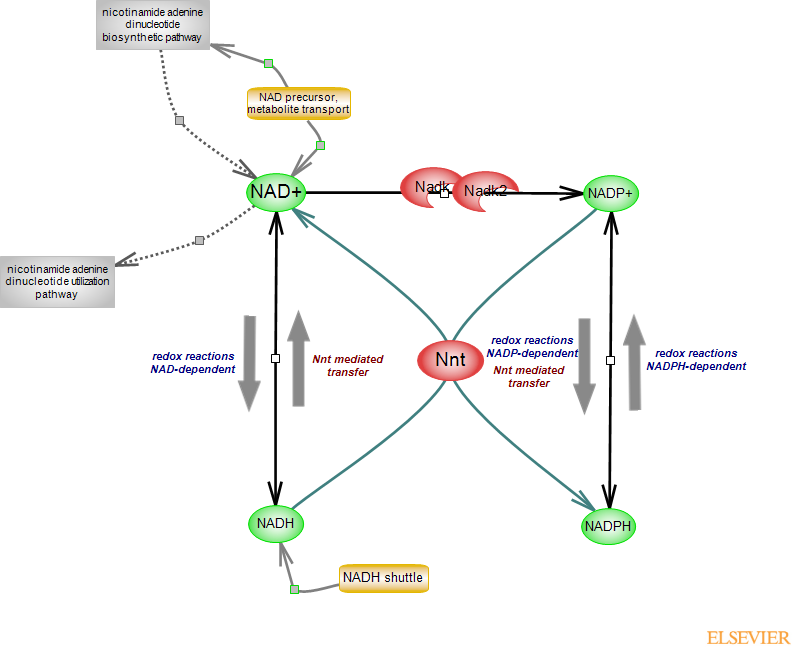
Glucose metabolism and reactions in other metabolic networks generate NADH from NAD. NADHs (and FADH2) formed during the tricarboxylic acid cycle will donate their electrons to fuel the electrochemical gradient the electron transport chain (ETC) generates which drives ATP synthesis. NADP can be formed via two routes: a de novo route whereby NAD is phosphorylated by NAD kinases, and via the reactions carried out by NADPH-dependent enzymes, converting the reduced molecule to its oxidized form. NADPH can also be formed in two ways: conversion of the oxidized molecule to its reduced form by NADP-dependent enzymes or via the reaction carried out by the mitochondrial transhydrogenase. Two enzymes in the pentose phosphate pathway, and the NADP-dependent cytosolic and mitochondrial isocitrate dehydrogenases and malic enzymes generate NADPH. The mitochondrial nicotinamide nucleotide transhydrogenase NNT carries out the conversion NADP + NADH <-> NAD + NADPH. As the reaction is proton-driven, the equilibrium is shifted far to the right, making the reaction almost unidirectional. The mitochondrial membrane is rather impermeable to NAD/NADH. The reducing equivalents of cytosolic NADH are shuttled into the mitochondria by NADH shuttles (redox shuttles): the malate-aspartate shuttle (MAS) and the glycerol-3-phosphate shuttle, and MAS plays the prominent role. Cytosolic and mitochondrial proteins compose the shuttles.
There are three pathways of NAD utilization: the sirtuin-mediated pathway and the ADP ribosylation pathways - mono and poly-ribosylation, and cyclic ribosylation. Sirtuins belong to class III deacetylases whose targets include histones and other proteins; a few could be involved in mono-ADP-ribosylation. There are 7 sirtuin proteins with various cellular localizations. Mono and poly-ADP-ribosylation are carried out by members of poly(ADP)-ribose polymerases - PARPs. There are 17 members in the mammalian family and apart from the highly-conserved ADP-ribosyl transferase (ART) catalytic domain show great structural domain variability. Most can only mono-ribosylate substrates (MAR), but a few can produce long chains (PAR), up to 200 units, and even introduce branching, occasionally. Cd38 and Cd157 are the enzymes involved in the formation of cyclic ADP-ribose (cADPR) and more is known about Cd38, although overall, less is known about this route of NAD consumption. cADPR prompts calcium release from intracellular stores. Another NAD derivative known to be a very potent calcium release agent is nicotinic acid adenine dinucleotide phosphate (NAADP), but the exact mode of its formation is rather debated.
To see the ontology report for GViewer, annotations and download, click here ...(less)
Pathway Diagram:
Genes in Pathway:
G
Gapdh
glyceraldehyde-3-phosphate dehydrogenase
ISO
SMPDB SMP:00124
NCBI chr 4:159,648,592...159,653,436
G
Got1
glutamic-oxaloacetic transaminase 1
ISO
RGD
PMID:18020963 PMID:16368075 RGD:5129954 , RGD:11251688
NCBI chr 1:252,306,541...252,337,622
G
Got2
glutamic-oxaloacetic transaminase 2
ISO
SMPDB PMID:18020963 PMID:16368075 SMP:00129, RGD:5129954 , RGD:11251688
NCBI chr19:9,180,428...9,206,113
G
Gpd1
glycerol-3-phosphate dehydrogenase 1
ISO
SMPDB PMID:18020963 PMID:16368075 SMP:00124, RGD:5129954 , RGD:11251688
NCBI chr 7:132,722,982...132,730,373
G
Gpd2
glycerol-3-phosphate dehydrogenase 2
ISO
SMPDB PMID:18020963 PMID:16368075 SMP:00124, RGD:5129954 , RGD:11251688
NCBI chr 3:62,209,432...62,346,590
G
Mdh1
malate dehydrogenase 1
ISO
RGD
PMID:18020963 PMID:16368075 RGD:5129954 , RGD:11251688
NCBI chr14:99,831,934...99,847,227
G
Mdh2
malate dehydrogenase 2
ISO
SMPDB PMID:18020963 PMID:16368075 SMP:00129, RGD:5129954 , RGD:11251688
NCBI chr12:26,530,886...26,543,841
G
Nadk
NAD kinase
ISO
RGD
PMID:25641397 RGD:11250407
NCBI chr 5:171,427,973...171,458,586
G
Nadk2
NAD kinase 2, mitochondrial
ISO
RGD
PMID:25641397 RGD:11250407
NCBI chr 2:59,844,854...59,886,989
G
Nnt
nicotinamide nucleotide transhydrogenase
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 2:53,144,157...53,237,857
G
Slc1a3
solute carrier family 1 member 3
ISO
RGD
PMID:18020963 PMID:16368075 RGD:5129954 , RGD:11251688
NCBI chr 2:59,482,707...59,557,808
G
Slc22a13
solute carrier family 22 member 13
ISO
RGD
PMID:25837229 RGD:11251687
NCBI chr 8:127,800,097...127,811,750
G
Slc25a11
solute carrier family 25 member 11
ISO
SMPDB SMP:00129
NCBI chr10:55,856,209...55,859,060
G
Slc25a12
solute carrier family 25 member 12
ISO
SMPDB PMID:18020963 PMID:16368075 SMP:00129, RGD:5129954 , RGD:11251688
NCBI chr 3:76,504,868...76,599,536
G
Slc25a13
solute carrier family 25 member 13
ISO
RGD
PMID:16368075 RGD:11251688
NCBI chr 4:35,145,721...35,328,403
G
Slc36a4
solute carrier family 36 member 4
ISO
RGD
PMID:25837229 RGD:11251687
NCBI chr 8:20,801,256...20,834,046
G
Slc5a8
solute carrier family 5 member 8
ISO
RGD
PMID:25837229 RGD:11251687
NCBI chr 7:25,160,280...25,201,185
G
Slc7a5
solute carrier family 7 member 5
ISO
RGD
PMID:25837229 RGD:11251687
NCBI chr19:66,843,808...66,872,412
G
Bst1
bone marrow stromal cell antigen 1
ISO
RGD
PMID:23576305 RGD:11250406
NCBI chr14:71,466,179...71,482,671
G
Cd38
CD38 molecule
ISO
RGD
PMID:23576305 RGD:11250406
NCBI chr14:71,384,532...71,424,794
G
Aplf
aprataxin and PNKP like factor
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 4:121,627,800...121,679,980
G
Chfr
checkpoint with forkhead and ring finger domains
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr12:52,164,214...52,197,860
G
Macrod1
mono-ADP ribosylhydrolase 1
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 1:213,675,413...213,831,571
G
Macrod2
mono-ADP ribosylhydrolase 2
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 3:148,173,821...150,191,077
G
Oard1
O-acyl-ADP-ribose deacylase 1
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 9:20,074,507...20,084,946
G
Parg
poly (ADP-ribose) glycohydrolase
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr16:7,442,696...7,550,585
G
Parp1
poly (ADP-ribose) polymerase 1
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr13:94,839,484...94,871,295
G
Parp10
poly (ADP-ribose) polymerase family, member 10
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 7:109,829,721...109,839,054
G
Parp11
poly (ADP-ribose) polymerase family, member 11
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 4:161,984,192...162,030,550
G
Parp12
poly (ADP-ribose) polymerase family, member 12
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 4:68,793,411...68,851,214
G
Parp14
poly (ADP-ribose) polymerase family, member 14
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr11:78,408,134...78,440,201
G
Parp16
poly (ADP-ribose) polymerase family, member 16
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 8:74,626,399...74,644,610
G
Parp2
poly (ADP-ribose) polymerase 2
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr15:26,494,722...26,517,893
G
Parp3
poly (ADP-ribose) polymerase family, member 3
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 8:115,989,721...115,995,822
G
Parp4
poly (ADP-ribose) polymerase family, member 4
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr15:34,806,203...34,908,492
G
Parp6
poly (ADP-ribose) polymerase family, member 6
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 8:68,912,393...68,944,904
G
Parp8
poly (ADP-ribose) polymerase family, member 8
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 2:50,408,404...50,581,513
G
Parp9
poly (ADP-ribose) polymerase family, member 9
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr11:78,286,282...78,320,409
G
Rnf146
ring finger protein 146
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 1:30,286,680...30,304,367
G
Tiparp
TCDD-inducible poly(ADP-ribose) polymerase
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 2:152,063,769...152,090,414
G
Tnks
tankyrase
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr16:63,922,969...64,073,972
G
Tnks2
tankyrase 2
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 1:243,980,432...244,033,629
G
Zc3hav1
zinc finger CCCH-type containing, antiviral 1
ISO
RGD
PMID:26091342 RGD:11100038
NCBI chr 4:67,979,055...68,029,353
G
Nadsyn1
NAD synthetase 1
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 1:208,410,914...208,439,242
G
Nmnat1
nicotinamide nucleotide adenylyltransferase 1
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 5:165,193,301...165,211,213
G
Nmnat2
nicotinamide nucleotide adenylyltransferase 2
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr13:67,655,794...67,831,609
G
Nmnat3
nicotinamide nucleotide adenylyltransferase 3
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 8:107,771,124...107,888,528
G
Qprt
quinolinate phosphoribosyltransferase
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 1:191,148,715...191,164,006
G
Nadsyn1
NAD synthetase 1
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 1:208,410,914...208,439,242
G
Nampt
nicotinamide phosphoribosyltransferase
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 6:55,152,756...55,189,547
G
Naprt
nicotinate phosphoribosyltransferase
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 7:109,457,328...109,460,817
G
Nmnat1
nicotinamide nucleotide adenylyltransferase 1
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 5:165,193,301...165,211,213
G
Nmnat2
nicotinamide nucleotide adenylyltransferase 2
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr13:67,655,794...67,831,609
G
Nmnat3
nicotinamide nucleotide adenylyltransferase 3
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 8:107,771,124...107,888,528
G
Nmrk1
nicotinamide riboside kinase 1
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 1:225,475,975...225,503,414
G
Nmrk2
nicotinamide riboside kinase 2
ISO
RGD
PMID:20007326 RGD:11099972
NCBI chr 7:9,165,567...9,168,638
G
Sirt1
sirtuin 1
ISO
RGD
PMID:26785480 RGD:11100032
NCBI chr20:25,305,953...25,328,000
G
Sirt2
sirtuin 2
ISO
RGD
PMID:26785480 RGD:11100032
NCBI chr 1:93,181,472...93,204,499
G
Sirt3
sirtuin 3
ISO
RGD
PMID:26785480 RGD:11100032
NCBI chr 1:205,371,703...205,394,145
G
Sirt4
sirtuin 4
ISO
RGD
PMID:26785480 RGD:11100032
NCBI chr12:46,785,852...46,800,179
G
Sirt5
sirtuin 5
ISO
RGD
PMID:26785480 RGD:11100032
NCBI chr17:21,515,982...21,543,529
G
Sirt6
sirtuin 6
ISO
RGD
PMID:26785480 RGD:11100032
NCBI chr 7:8,733,056...8,738,543
G
Sirt7
sirtuin 7
ISO
RGD
PMID:26785480 RGD:11100032
NCBI chr10:106,394,802...106,401,627
Pathway Gene Annotations
Disease Annotations Associated with Genes in the nicotinamide adenine dinucleotide metabolic pathway
Gapdh Acute Coronary Syndrome , Alzheimer's disease , Animal Disease Models , brain glioma , Colorectal Neoplasms , COVID-19 , diabetic retinopathy , epilepsy , Esophageal Neoplasms , Experimental Diabetes Mellitus , hepatocellular carcinoma , Huntington's disease , Hypoxia , lung adenocarcinoma , lymphangioleiomyomatosis , middle cerebral artery infarction , Mouth Neoplasms , Myocardial Reperfusion Injury , Necrosis , obesity , oral squamous cell carcinoma , osteoarthritis , osteoporosis , Parkinsonism , Pregnancy in Diabetics , rheumatic heart disease , Spinal Cord Injuries , Spinal Cord Reperfusion Injury , squamous cell carcinoma , type 2 diabetes mellitus , Viral Bronchiolitis Got1 Acute Lung Injury , amyloidosis , amyotrophic lateral sclerosis , cardiomyopathy , Chemical and Drug Induced Liver Injury , Closed Head Injuries , Experimental Liver Cirrhosis , Febrile Seizures , Hyperoxia , pancreatic ductal adenocarcinoma , Pulmonary Hypertension, Hypoxia-Induced , transient cerebral ischemia Got2 acute kidney failure , developmental and epileptic encephalopathy , developmental and epileptic encephalopathy 82 , early infantile epileptic encephalopathy , Febrile Seizures , neurodegenerative disease , obesity , Pulmonary Hypertension, Hypoxia-Induced , schizophrenia , Spinal Cord Injuries , Transplant Rejection Gpd1 Chemical and Drug Induced Liver Injury , genetic disease , Hypertriglyceridemia, Transient Infantile Gpd2 COVID-19 , Experimental Diabetes Mellitus , maturity-onset diabetes of the young type 1 , methylmalonic acidemia , osteoporosis , type 2 diabetes mellitus Mdh1 Acute Liver Failure , adult respiratory distress syndrome , Chemical and Drug Induced Liver Injury , developmental and epileptic encephalopathy 88 , Experimental Liver Cirrhosis , hypertension , Liver Injury Mdh2 Animal Mammary Neoplasms , brain disease , brain ischemia , carcinoma , congestive heart failure , developmental and epileptic encephalopathy 1 , developmental and epileptic encephalopathy 51 , Experimental Mammary Neoplasms , genetic disease , myocardial infarction , ovarian carcinoma Nadk2 2,4-Dienoyl-CoA Reductase Deficiency , genetic disease Nnt genetic disease , glucocorticoid deficiency 1 , Glucocorticoid Deficiency 4 , obesity Slc1a3 autistic disorder , COVID-19 , epilepsy , episodic ataxia , episodic ataxia type 6 , Experimental Diabetes Mellitus , genetic disease , low tension glaucoma , spastic ataxia Slc25a11 adult respiratory distress syndrome , pheochromocytoma/paraganglioma syndrome 6 Slc25a12 Asperger syndrome , autism spectrum disorder , autistic disorder , developmental and epileptic encephalopathy 1 , developmental and epileptic encephalopathy 39 , fetal alcohol spectrum disorder , genetic disease , rheumatoid arthritis Slc25a13 adult-onset type II citrullinemia , Bone Fractures , citrullinemia , Citrullinemia Type 2 , classic citrullinemia , Experimental Liver Cirrhosis , genetic disease , hereditary spastic paraplegia 11 , megacolon , neonatal-onset type II citrullinemia Slc5a8 Colonic Neoplasms , hyperuricemia Slc7a5 adenoid cystic carcinoma , allergic contact dermatitis , autism spectrum disorder , bile duct adenocarcinoma , bile duct carcinoma , Chemical and Drug Induced Liver Injury , cholangiocarcinoma , colon cancer , colorectal adenocarcinoma , colorectal cancer , Disease Progression , Endotoxemia , esophageal carcinoma , esophagitis , esophagus squamous cell carcinoma , Experimental Liver Neoplasms , extrahepatic bile duct adenocarcinoma , gastric adenocarcinoma , glioblastoma , hepatocellular carcinoma , high grade glioma , large cell neuroendocrine carcinoma , Liver Metastasis , lung adenocarcinoma , Lung Neoplasms , lung non-small cell carcinoma , lung pleomorphic carcinoma , lung squamous cell carcinoma , Lymphatic Metastasis , malignant astrocytoma , malignant pleural mesothelioma , Neoplasm Metastasis , oral squamous cell carcinoma , rectum adenocarcinoma , Right Ventricular Hypertrophy , Spinal Cord Injuries , stomach cancer , tongue squamous cell carcinoma , transitional cell carcinoma
2,4-Dienoyl-CoA Reductase Deficiency Nadk2 Acute Coronary Syndrome Gapdh acute kidney failure Got2 Acute Liver Failure Mdh1 Acute Lung Injury Got1 adenoid cystic carcinoma Slc7a5 adult respiratory distress syndrome Mdh1 , Slc25a11 adult-onset type II citrullinemia Slc25a13 allergic contact dermatitis Slc7a5 Alzheimer's disease Gapdh amyloidosis Got1 amyotrophic lateral sclerosis Got1 Animal Disease Models Gapdh Animal Mammary Neoplasms Mdh2 Asperger syndrome Slc25a12 autism spectrum disorder Slc25a12 , Slc7a5 autistic disorder Slc1a3 , Slc25a12 bile duct adenocarcinoma Slc7a5 bile duct carcinoma Slc7a5 Bone Fractures Slc25a13 brain disease Mdh2 brain glioma Gapdh brain ischemia Mdh2 carcinoma Mdh2 cardiomyopathy Got1 Chemical and Drug Induced Liver Injury Got1 , Gpd1 , Mdh1 , Slc7a5 cholangiocarcinoma Slc7a5 citrullinemia Slc25a13 Citrullinemia Type 2 Slc25a13 classic citrullinemia Slc25a13 Closed Head Injuries Got1 colon cancer Slc7a5 Colonic Neoplasms Slc5a8 colorectal adenocarcinoma Slc7a5 colorectal cancer Slc7a5 Colorectal Neoplasms Gapdh congestive heart failure Mdh2 COVID-19 Gapdh , Gpd2 , Slc1a3 developmental and epileptic encephalopathy Got2 developmental and epileptic encephalopathy 1 Mdh2 , Slc25a12 developmental and epileptic encephalopathy 39 Slc25a12 developmental and epileptic encephalopathy 51 Mdh2 developmental and epileptic encephalopathy 82 Got2 developmental and epileptic encephalopathy 88 Mdh1 diabetic retinopathy Gapdh Disease Progression Slc7a5 early infantile epileptic encephalopathy Got2 Endotoxemia Slc7a5 epilepsy Gapdh , Slc1a3 episodic ataxia Slc1a3 episodic ataxia type 6 Slc1a3 esophageal carcinoma Slc7a5 Esophageal Neoplasms Gapdh esophagitis Slc7a5 esophagus squamous cell carcinoma Slc7a5 Experimental Diabetes Mellitus Gapdh , Gpd2 , Slc1a3 Experimental Liver Cirrhosis Got1 , Mdh1 , Slc25a13 Experimental Liver Neoplasms Slc7a5 Experimental Mammary Neoplasms Mdh2 extrahepatic bile duct adenocarcinoma Slc7a5 Febrile Seizures Got1 , Got2 fetal alcohol spectrum disorder Slc25a12 gastric adenocarcinoma Slc7a5 genetic disease Gpd1 , Mdh2 , Nadk2 , Nnt , Slc1a3 , Slc25a12 , Slc25a13 glioblastoma Slc7a5 glucocorticoid deficiency 1 Nnt Glucocorticoid Deficiency 4 Nnt hepatocellular carcinoma Gapdh , Slc7a5 hereditary spastic paraplegia 11 Slc25a13 high grade glioma Slc7a5 Huntington's disease Gapdh Hyperoxia Got1 hypertension Mdh1 Hypertriglyceridemia, Transient Infantile Gpd1 hyperuricemia Slc5a8 Hypoxia Gapdh large cell neuroendocrine carcinoma Slc7a5 Liver Injury Mdh1 Liver Metastasis Slc7a5 low tension glaucoma Slc1a3 lung adenocarcinoma Gapdh , Slc7a5 Lung Neoplasms Slc7a5 lung non-small cell carcinoma Slc7a5 lung pleomorphic carcinoma Slc7a5 lung squamous cell carcinoma Slc7a5 lymphangioleiomyomatosis Gapdh Lymphatic Metastasis Slc7a5 malignant astrocytoma Slc7a5 malignant pleural mesothelioma Slc7a5 maturity-onset diabetes of the young type 1 Gpd2 megacolon Slc25a13 methylmalonic acidemia Gpd2 middle cerebral artery infarction Gapdh Mouth Neoplasms Gapdh myocardial infarction Mdh2 Myocardial Reperfusion Injury Gapdh Necrosis Gapdh neonatal-onset type II citrullinemia Slc25a13 Neoplasm Metastasis Slc7a5 neurodegenerative disease Got2 obesity Gapdh , Got2 , Nnt oral squamous cell carcinoma Gapdh , Slc7a5 osteoarthritis Gapdh osteoporosis Gapdh , Gpd2 ovarian carcinoma Mdh2 pancreatic ductal adenocarcinoma Got1 Parkinsonism Gapdh pheochromocytoma/paraganglioma syndrome 6 Slc25a11 Pregnancy in Diabetics Gapdh Pulmonary Hypertension, Hypoxia-Induced Got1 , Got2 rectum adenocarcinoma Slc7a5 rheumatic heart disease Gapdh rheumatoid arthritis Slc25a12 Right Ventricular Hypertrophy Slc7a5 schizophrenia Got2 spastic ataxia Slc1a3 Spinal Cord Injuries Gapdh , Got2 , Slc7a5 Spinal Cord Reperfusion Injury Gapdh squamous cell carcinoma Gapdh stomach cancer Slc7a5 tongue squamous cell carcinoma Slc7a5 transient cerebral ischemia Got1 transitional cell carcinoma Slc7a5 Transplant Rejection Got2 type 2 diabetes mellitus Gapdh , Gpd2 Viral Bronchiolitis Gapdh