Gene Expression and Regulation Pathway Suite Network
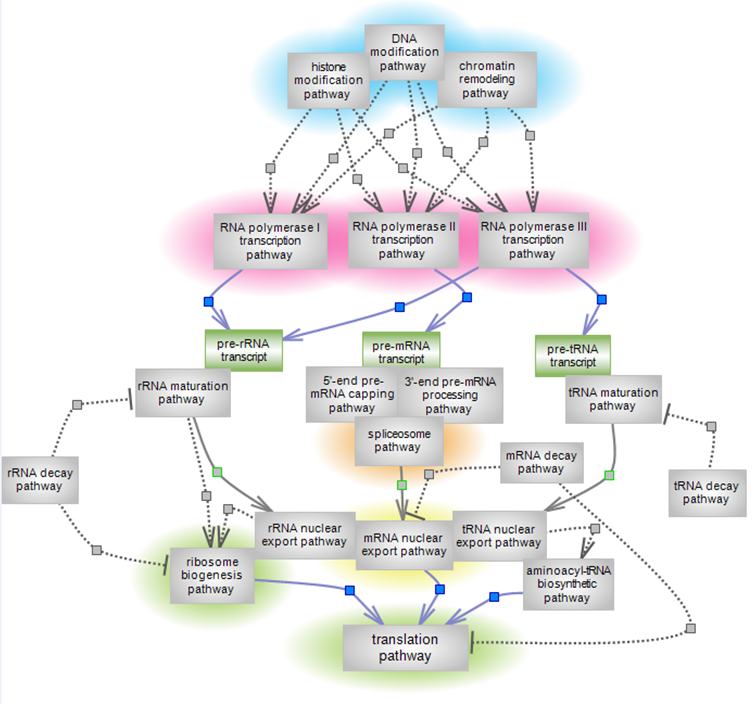 |
The expression of eukaryotic genes into functional RNA or protein molecules is dependent upon the activity of three polymerases and accompanying processing, transport and surveillance systems that forge and traffic the nascent transcripts, assure the proper quality and delivery of the final products. Differential gene expression – which gene is expressed when and where – is directly affected by the relative relaxation or compaction of chromatin, which also impacts on DNA replication, DNA damage response and repair, and associated RNA processing. Epigenetic changes, the modification of DNA and histone and the remodeling of chromatin, are thus key to the spatiotemporal expression of genes and related events. Chromatin modification involves the methylation of DNA and the many ways in which histone tails can be modified. The modification pathways bring together the enzymes that catalyze the addition or removal of modifications, known as ‘writers’ and ‘erasers’, respectively, and the proteins whose particular domains recognize the modification, the ‘readers’. The remodeling of chromatin involves the sliding, spacing or repositioning of entire nucleosomes, and the insertion, eviction or exchange of histones. The assembly and disassembly of chromatin is ATP-dependent and is carried out by four protein complexes in four remodeling pathways with distinct functions. The pathways of epigenetic changes are highlighted in blue. The three eukaryotic RNA polymerases (RNAPs) – Pol I, II and III, transcribe different sets of genes and require distinct sets of factors to assemble into a specific initiation complex that recruits them on gene promoters. All share an initiation, elongation and termination step, but they differ in detail and/or extent of complexity in the three systems. Although representing a small percentage of the genome, the expression of proteins engages all three polymerases – pol II, which transcribes the mRNA of protein-coding genes, pol I, which transcribes the ribosomal genes (rRNA) and pol III, which transcribes the transfer RNA (tRNA) and also one of the four rRNA genes. The three transcription pathways are highlighted in red; the spliceosome pathway is highlighted in orange. |
| Intimately connected to transcription are many of the pre-transcript maturation and nuclear export events. Particularly in the case of mRNA, 5’ pre-mRNA and 3’ pre-mRNA processing are coupled to initiation of transcription elongation and termination, respectively. Most mRNAs contain introns whose splicing is largely co-transcriptional and for which the end of one splicing cycle couples to the export of mature mRNA. tRNA and rRNA maturation occur both in the nucleus/nucleolus and in the cytoplasm. rRNA maturation, the production of the four rRNA genes, is coupled to the generation of the pre-ribosomal 40S and 60S particles. The particles are exported to the cytoplasm where the final steps of ribosome biogenesis yield the translation-competent ribosomes. Along the way, surveillance mechanisms are in place for quality control (QC) of the correctness of RNA molecules and initiate decay or recycling events, as necessary. Completion of ribosome biogenesis and aminoacylation of tRNA fuel the translation of mRNA into a peptide chain followed by its folding into the protein structure, protein post-translational modification and delivery to site(s) of action. The spliceosome pathway is highlighted in orange; the pathway of mRNA nuclear export is highlighted in yellow; the ribosome biogenesis and translation pathways are highlighted in green. |
|
Click here to explore the Epigenetic Regulation/Control – Chromatin Modification/Remodeling Pathway Suite.
Click here to explore the Transcription and Transcription-Coupled Events Pathway Suite.
Click here to explore the RNA Maturation, Transport and Surveillance (QC) and Protein Translation Pathway Suite.
Epigenetic Regulation/Control – Chromatin Modification/Remodeling Pathway Suite
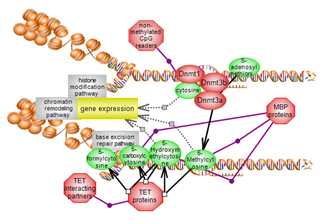
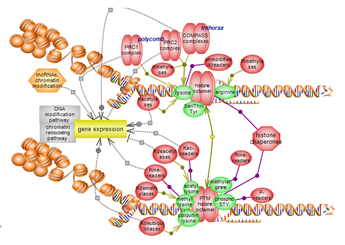
The relative compaction or relaxation of chromatin impacts on gene transcription and associated events. This suite brings together the pathways of DNA and histone modification and of chromatin remodeling. The pathways are highlighted in blue.
| Chromatin remodeling pathway | DNA modification pathway | Histone modification pathway |
| Chromatin remodeling is carried out by four major protein complexes representing four remodeling pathways with distinct functions. The four major families/complexes of ATP-dependent chromatin remodelers are: SWI/SNF, INO80, ISWI and CHD. Each contains specific sub-complexes to carry out the many aspects of chromatin remodeling. The complexes are collectively shown in the diagram and individually described. Click here to explore this intricate epigenetic pathway. |
The DNA modification pathway involves the methylation of cytosines within CpG genomic dinucleotides. Methylated and non-methylated CpGs are binding partners for numerous players. Members of the TET family of dioxygenases act upon the methylated cysteine and are also binding partners. A complex interplay exists between DNA methylation and histone modification, particularly histone lysine methylation. Click here to explore this important epigenetic pathway. |
The histone modification pathway involves several residues, particularly lysines, and numerous modification types. Many classes of enzymes are involved in the various modification types while others act to remove them. Several domain types participate in the recognition of modified residues with unmodified also acting as binding partners. A complex interplay exists between histone modification, particularly histone lysine methylation, and DNA methylation. Click here to explore several aspects of this very important epigenetic pathway. |
Return to pathway suite network diagram
Transcription and Transcription-Coupled Events Pathway Suite
Eukaryotic transcription is carried out by three polymerase systems. In the case of mRNA, the maturation of the pre-mRNA transcript is coupled to transcription. This suite brings together the transcription pathways of RNA polymerases I, II and III and the spliceosome pathway, part of mRNA maturation that largely occurs co-transcriptionally. The transcription pathways are highlighted in red; the spliceosome pathway, also in another suite, is highlighted in orange.
| RNA polymerase I transcription pathway | RNA polymerase II transcription pathway |
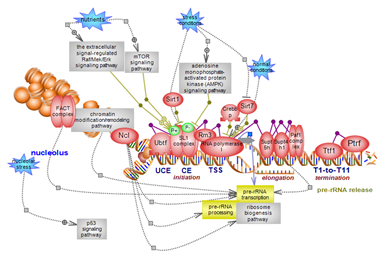
| Nucleolar RNA polymerase I transcription has one gene as its target: the large 47S ribosomal gene whose transcript is further processed into the three mature 18S, 28S and 5.8S rRNAs. Multiple copies of the gene are distributed in clusters on several chromosomes. As part of the highly energy-consuming process of ribosome biosynthesis, the rRNA transcription pathway is highly regulated at multiple levels. Click here to explore this important regulatory pathway. |
The three mammalian RNA polymerases transcribe distinct classes of genes. RNA polymerase II (RNAPII or pol II) transcribes all protein-coding genes and many of the non-coding genes. With the genome being pervasively transcribed, pol II is one of the busiest transcription systems, also the most complex. The carboxyl-terminal domain (CTD) of the largest subunit of pol II is unique to this polymerase and plays major roles in the elaborate initiation, elongation and termination steps as well as associated processes. The Mediator and other specific complexes fine-tune the regulation of the system and its coupling with co-transcriptional events such as splicing. These are among the salient features of the pol II transcription pathway. Click here to explore this intricate system. |
| RNA polymerase III transcription pathway | Spliceosomal pathway |
| The three mammalian RNA polymerases transcribe distinct classes of genes. RNA polymerase III (RNAPIII or pol III) transcribes all transfer RNA (tRNA) genes, the small ribosomal 5S (5srRNA) and several small, non-coding RNA genes, including the U6 spliceosomal RNA. Click here to explore this important transcriptional pathway. |
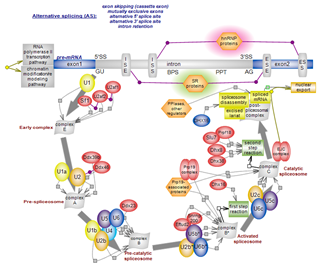
The spliceosome – the multimegadalton ribonucleoprotein complex, is one of the most elaborate macromolecular machines of the cell. Five small nuclear ribonucleoprotein particles (snRNPs) assemble in discrete complexes along a dynamic, energy-fueled pathway. Substantial structural and functional rearrangements along this route promote the catalytically competent spliceosomes that perform the two esterification reactions that excise intervening introns and ligate the exons of pre-mRNAs transcribed by RNA polymerase II. Splicing is largely co-transcriptional, and most genes in higher eukaryotes are subject to alternative splicing (AS). Alternative exons can be included or skipped, alternative 5’ and 3’ splice sites can be used and certain introns can be retained. AS vastly expands the repertoire of proteins and the range of their functional versatility. Chromatin dynamics and structure impact on transcription, splicing and alternative splicing. Splicing, in turn, affects transcriptional rates and the modulation of chromatin configuration. The pathways of transcription, splicing and chromatin modification and remodeling are entwined in a continuous, bidirectional partnership. Click here to explore the intricate spliceosomal pathway. |
Return to pathway suite network diagram
RNA Maturation, Transport and Surveillance (QC) and Protein Translation Pathway Suite
RNA processing into a mature, functional form, nuclear export and in the case of rRNA, assembly of translation-competent ribosomes, along with surveillance mechanisms, assure the quality of the final step of peptide biosynthesis. This suite brings together the spliceosome pathway (also shown in the ‘Transcription and Transcription-Coupled Events Pathway Suite’), highlighted in orange, the pathway of mRNA nuclear export, highlighted in yellow and the ribosome biogenesis and translation pathways, highlighted in green.
| Spliceosomal pathway | mRNA nuclear export pathway |
| The spliceosome – the multimegadalton ribonucleoprotein complex, is one of the most elaborate macromolecular machines of the cell. Five small nuclear ribonucleoprotein particles (snRNPs) assemble in discrete complexes along a dynamic, energy-fueled pathway. Substantial structural and functional rearrangements along this route promote the catalytically competent spliceosomes that perform the two esterification reactions that excise intervening introns and ligate the exons of pre-mRNAs transcribed by RNA polymerase II. Splicing is largely co-transcriptional, and most genes in higher eukaryotes are subject to alternative splicing (AS). Alternative exons can be included or skipped, alternative 5’ and 3’ splice sites can be used and certain introns can be retained. AS vastly expands the repertoire of proteins and the range of their functional versatility. Chromatin dynamics and structure impact on transcription, splicing and alternative splicing. Splicing, in turn, affects transcriptional rates and the modulation of chromatin configuration. The pathways of transcription, splicing and chromatin modification and remodeling are entwined in a continuous, bidirectional partnership. Click here to explore the intricate spliceosomal pathway. |
Mature mRNA that has been 5’-capped, spliced and 3’-polyadenylated has to be exported to the cytoplasm to serve as the template for protein biosynthesis. Its transport through the nuclear pore complex (NPC) is an important element of regulated gene expression. Nuclear export is dependent upon transport receptors and the adaptors that couple them to cargo. Click here to explore this important regulatory pathway. |
| Ribosome biogenesis pathway | Translation pathway |
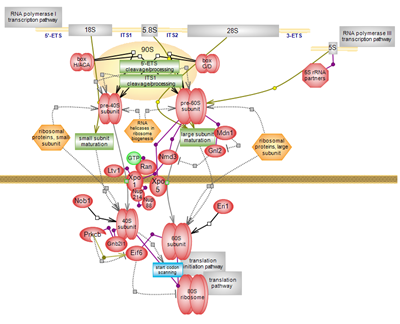
| Ribosome biogenesis is an essential process and is one of the most energy-consuming cellular events. It plays a central role in gene expression as the translationally-competent ribosome is involved in decoding the genetic information encrypted in mRNA into the polypeptide chain of functional proteins. In all organisms, the ribosome is built of two subunits: the small, 40S subunit in eukaryotes that acts as the mRNA decoder and the large 60S eukaryotic subunit that carries out the peptidyl transferase reaction. The complex assembly journey starts in the nucleolus/nucleus and ends in the cytoplasm, brings the ribosomal proteins specific for each subunit together and includes some 200 plus non-ribosomal, trans-acting factors involved or assisting in the folding and modification, cleavage and processing of rRNA. Click here to explore the details of the fascinating odyssey of ribosome biogenesis. |
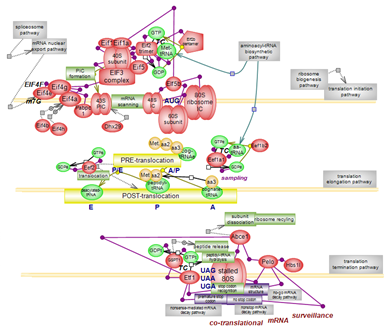
Translation is the final stage of protein-coding gene expression. The genetic information encoded in the mature mRNA transcript is decoded into the polypeptide chain by the ribosome, a massive 80S ribonucleoprotein complex whose small 40S subunit is the mRNA decoder and the large 60S subunit carries out the peptidyl transferase reaction. Three steps are involved in the task: initiation, elongation and termination followed by the splitting of ribosomes and their recycling. Of these, the initiation pathway is the rate-limiting and the most regulated step. Initiation is intimately connected to the biogenesis of ribosomes as joining of the 60S subunit and formation of the elongation-competent 80S ribosome is subsequent to finding the initiation AUG codon by the methionine-tRNA-loaded preinitiation complex (PIC). Along the route, reactions and interactions, GTP hydrolysis and conformational changes mold the unfolding of translation pathways. Co-translational mRNA surveillance mechanisms are in place to rescue/recycle ribosomes aberrantly stalled due to structural features of mRNA, the presence of a premature stop codon or its absence in the mRNA sequence. Click here to explore the details of this complex system. |
Return to pathway suite network diagram