Parkinson Disease Pathway Suite
|
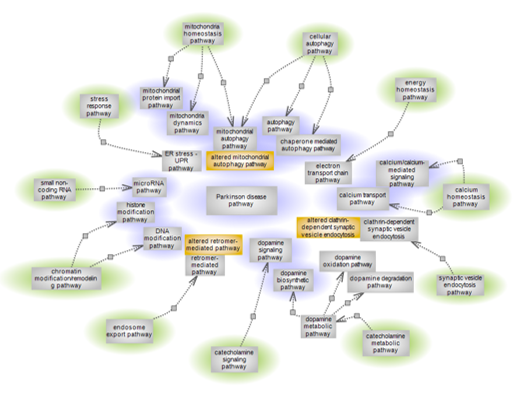
| Parkinson’s disease (PD) is a progressive neurodegenerative condition of complex etiology exhibiting a range of movement syndromes caused by the selective degeneration of dopaminergic neurons in the substantia nigra pars compacta (SNc). It is the second most prevalent neurodegenerative disease and largely sporadic, with age representing a major risk factor. Several culprit genes and associated pathways play roles in the condition while other PD genes are still to be functionally characterized. To give a global view of pathway categories and individual pathways within that in some fashion relate to PD, a pathway suite is being offered. The pathway categories are highlighted in green and individual pathways within each category that are of relevance to, or affected by the condition are shown. Highlighted in purple are the pathways that currently have interactive diagram pages. Altered pathways are shown color-coded in orange. The purple highlighted pathways by the category/categories to which they belong are listed below with brief description and links to individual diagram pages. |
| Click title to go to: |
Parkinson disease pathway
Mitochondria homeostasis pathways
Cellular autophagy pathways
Chromatin modification/remodeling pathways and small non-coding RNA pathway
Calcium homeostasis pathways
Catecholamine metabolic and signaling pathways
Other Suites and Suite Networks
|
Parkinson disease pathway
|
|
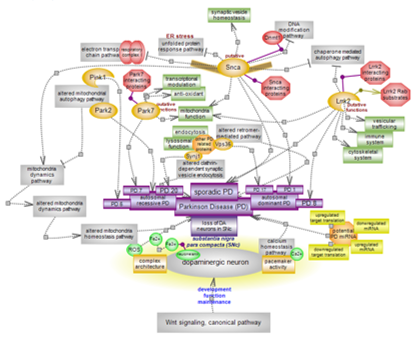
| Parkinson’s disease (PD) is a progressive neurodegenerative condition of complex etiology exhibiting a range of movement syndromes caused by the selective degeneration of dopaminergic neurons in the substantia nigra pars compacta (SNc). It is the second most prevalent neurodegenerative disease and is largely sporadic, with age representing a major risk factor. Several culprit genes and associated pathways play roles in the condition while other PD genes are still to be functionally characterized. Mitochondrial function declines with age. Alterations in mitochondrial function and homeostasis and alterations in other, related or important pathways can contribute to PD unfolding. Effects are likely augmented by the unique features of dopaminergic neurons in this brain area. Click here to explore aspects of this still enigmatic system.
Return to overview |
Mitochondria homeostasis pathways
Mitochondrial autophagy pathway
 |
Altered mitochondrial autophagy pathway

|
|
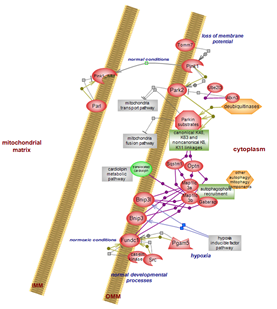
Removal of mitochondria – mitochondrial autophagy or mitophagy –
proceeds via alternative routes. One involves the Pink1-Parkin duo responding
to loss of mitochondrial membrane potential and mediated via ubiquitination
events. Another route involves the mitophagy receptors and responds to stresses
such as hypoxia or facilitates mitochondria removal under normal physiological,
developmental conditions. Click here
to explore the details of this complex system.
|
Mitochondrial autophagy (mitophagy) is a specialized aspect of the
overall, non-selective bulk macroautophagy, or autophagy pathway. Pink1-Parkin
is the main route of mitochondria quality control (QC) involving the
nuclear-encoded mitochondrial serine/threonine kinase Pink1 and downstream of
it, the cytosolic E3 ubiquitin ligase Parkin (Park2). Inactivating mutations in
the two proteins impair mitochondrial autophagy and also negatively impact on
mitochondria fusion and transport by abrogating the inhibitory but protective
effect they exert. Click here
to examine the details.
|
Mitochondria dynamics pathway
 |
Mitochondrial protein import pathway
 |
|
Mitochondrial dynamics consists of fission, fusion and transport
pathways. Fission allows for segregation of mitochondria with healthy
membrane potential from those with less than optimal membranes. It also
promotes mitochondrial autophagy. Fusion allows for the formation of
mitochondria networks. transport assures the delivery of mitochondria to places
of energy demand, of which the neural synapse have some of the highest. Both
fusion and transport can be inhibited by mitophagy. Click here
to explore this important network system.
Return to overview
|
Mitochondria perform essential cellular functions. In addition to the major function of ATP production, mitochondria are the site of fatty acid oxidation, the citric acid cycle and urea cycle, and heme and iron-sulfur cluster biosynthesis. They also provide for calcium storage and signaling and have a central role in apoptosis. Yet, only ~1% of mitochondrial proteins are encoded in the mitochondrial DNA; the remaining ~99% nuclear-encoded proteins have to be imported. As such, the mitochondrial protein import pathway impacts on every aspect of mitochondrial function. Click here to explore this complex system of five specialized import pathways. |
Cellular autophagy pathways
Autophagy pathway
 |
Mitochondrial autophagy pathway
 |
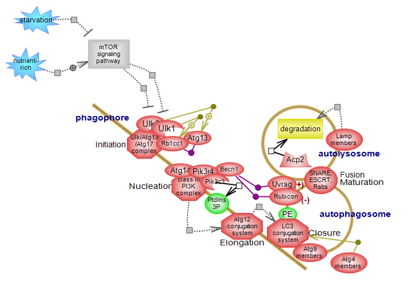
| Autophagy pathway (the macroautophagy arm of cellular autophagy) plays essential roles in cellular and tissues homeostasis and it acts as a source of nutrients under starvation or stress conditions. It involves the sequestration of damaged organelles, long-lived or aggregated proteins within double membrane structures and the subsequent delivery of cargo to lysosomes for degradation. Autophagy proceeds via several steps whose main components have been identified, although the molecular details and underlying mechanisms are still to be determined. Click here to explore this very important regulatory system. |
Removal of mitochondria – mitochondrial autophagy or mitophagy –
proceeds via alternative routes. One involves the Pink1-Parkin duo responding
to loss of mitochondrial membrane potential and mediated via ubiquitination
events. Another route involves the mitophagy receptors and responds to stresses
such as hypoxia or facilitates mitochondria removal under normal physiological,
developmental conditions. Click here
to explore the details of this complex system.
|
Chaperone mediated autophagy pathway
|
Altered mitochondrial autophagy pathway
 |
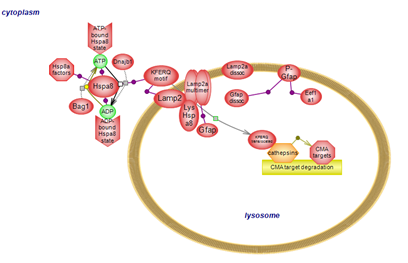
| Chaperone mediated autophagy (CMA) is a selective autophagy pathway whereby cytosolic proteins bearing a recognition motif are individually recognized and translocated into the lysosomal lumen for degradation. Targets of CMA include proteins associated with PD; pathogenic mutations in these genes affect their processing by CMA. Click here to explore the details of this form of autophagy.
Return to overview |
Mitochondrial autophagy (mitophagy) is a specialized aspect of the overall, non-selective bulk macroautophagy, or autophagy pathway. Pink1-Parkin is the main route of mitochondria quality control (QC) involving the nuclear-encoded mitochondrial serine/threonine kinase Pink1 and downstream of it, the cytosolic E3 ubiquitin ligase Parkin (Park2). Inactivating mutations in the two proteins impair mitochondrial autophagy and also negatively impact on mitochondria fusion and transport by abrogating the inhibitory but protective effect they exert. Click here to examine the details. |
Chromatin modification/remodeling pathways and small non-coding RNA pathway
DNA modification pathway
 |
Histone modification pathway
 |
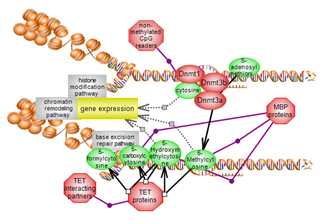
The DNA modification pathway involves the methylation of cytosines
within CpG genomic dinucleotides. Methylated and non-methylated CpGs are
binding partners for numerous players. Members of the TET family of
dioxygenases act upon the methylated cysteine and are also binding
partners. A complex interplay exists between DNA methylation and histone
modification, particularly histone lysine methylation. Click here to explore this important epigenetic pathway. |
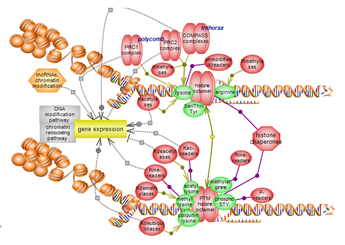
The histone modification pathway involves several residues, particularly
lysines, and numerous modification types. Many classes of enzymes are
involved in the various modification types while others act to remove
them. Several domain types participate in the recognition of modified
residues with unmodified also acting as binding partners. A complex
interplay exists between histone modification, particularly histone
lysine methylation, and DNA methylation. Click here to explore several aspects of this very important epigenetic pathway. |
microRNA pathway
 |
|
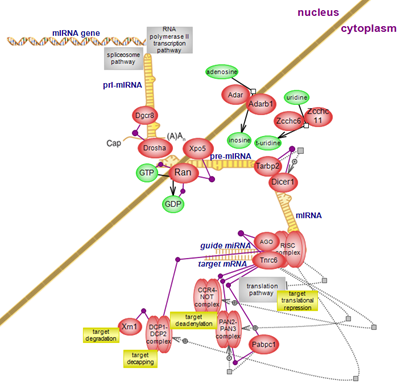
microRNAs are important regulators of gene expression via repression of
protein translation at the initiation and post-initiation steps and or by
inducing the deadenylation of target genes, followed by their decapping and
degradation. While the molecular mechanisms of translation inhibition are
poorly understood, the steps leading to the 5’-3’ degradation of target genes
are better documented. Click here to
explore the details of microRNA biogenesis and how the pathway impacts on the
stability of target genes.
Return to overview
|
Calcium homeostasis pathways
|
|
|
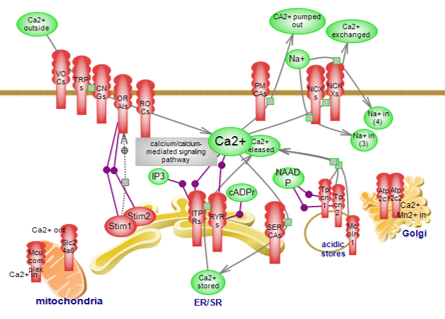
| Calcium transport pathway
|
|
Calcium/calcium-mediated signaling pathway

|
|
A wide array of channels, pumps, receptors and exchangers make
available or remove, as necessary, the Ca2+ ion, one of the most versatile
signaling molecules in living systems. Click here to explore
the complex pathway of Ca2+ transport.
Return to overview
|
|
The providers/regulators of the signal, exemplified in the calcium
transport pathway along with the buffers and sensors that further shape the
availability of the ion and its messaging, represent the components of the
‘calcium signaling kit’, and are presented in this overall diagram. Click
here to explore this complex circuit. |
Catecholamine metabolic and signaling pathways
|
|
|
|
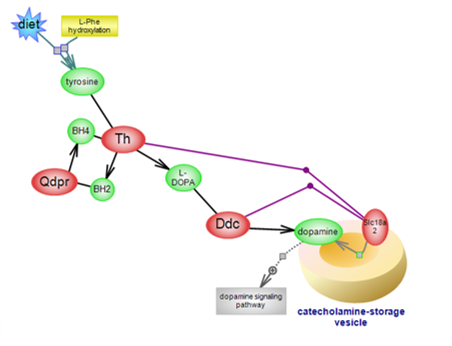
Dopamine biosynthetic pathway
|
|
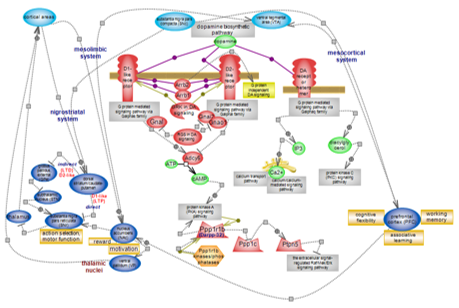
Dopamine signaling pathway
 |
|
Dopamine (DA) is the first of the three catecholamines whose synthesis
begins with L-tyrosine, derived from diet or from the hydroxylation of
L-phenylalanine. DA acts as a neurotransmitter and neuromodulator in the
brain and peripherally, as an autocrine/paracrine hormone. Click here to see the details of the biosynthetic pathway of this important molecule.
Return to overview
|
|
Dopamine (DA) exerts its major role in the nervous system where it is involved in motor control and action selection, reward and motivation, behavior, memory and cognition. The many effects are achieved through the projections dopaminergic (DA-producing) neurons establish and the inputs they receive which shape DA release. DA neurons in midbrain areas such as the substantia nigra pars compacta (SNc) and the ventral tegmental area (VTA), project to the striatum or the cerebral cortex to form specific neural pathways. Projections from the SNc to the dorsal striatum (caudate-putamen in primates) form the nigrostriatal system; from the VTA to the nucleus accumbens (NAc) and from the VTA (and SNc in primates) to the cortex, in particular the frontal lobes (prefrontal cortex, PFC), form the mesolimbic and the mesocortical system, respectively, sometimes collectively referred to as the mesocorticolimbic system. These are the three main DA systems. DA signals through five G protein-coupled receptors that are subdivided into two types, selectively expressed in target neurons. The role DA plays is largely modulatory. Click here to explore DA signaling and the systems it modulates. |
|







































