S-adenosylmethionine Homeostasis Pathway Suite Network
|
||
| S-adenosylmethionine (SAM) is a universal methyl donor and is at the cross roads of metabolism, gene expression regulation and cellular signaling. It is generated in the first reaction of the methionine cycle. The methylation of substrates will result in S-adenosylhomocysteine, the precursor of homocysteine, itself at the crossroads of several pathways. The remethylation of homocysteine will regenerate methionine and the major route is dependent upon the folate metabolic pathway. Also, homocysteine can proceed via the transsulfuration pathway to give rise to cysteine and derivatives. The metabolic pathways/cycles of methionine, homocysteine and folate are intimately connected and dependent upon each other. In the salvage pathway of methionine metabolism, SAM processing leads to the synthesis of polyamines. SAM is also a substrate for radical-SAM (RS) enzymes whose iron-sulfur cluster is used to generate the adenosyl radical. The intermediate radical allows these enzymes to mediate a vast range of chemical transformations important for co-factor and lipid metabolism, peptide and RNA modification. The methyltransferases are involved in the methylation of DNA and histones and play an important role in the epigenetic control of gene expression. The methylation of non-histone proteins and other molecules modulates the outcomes of cellular signaling. The pathway suite network of SAM homeostasis brings together elements from the suite dedicated to methionine, homocysteine, folate and related metabolites, and the suite network of gene expression and regulation along with other pertinent aspects. Highlighted are the pathways with currently available interactive diagram pages. |
Click here to explore the pathway suite for SAM homeostasis-related Metabolic Pathways.
Click here to explore the pathway suite for SAM homeostasis related Regulatory Pathways.
SAM-homeostasis-related Metabolic Pathway Suite
| Methionine cycle/metabolic pathway | Homocysteine metabolic pathway |
| The methionine cycle, via the de novo arm, produces the primary methyl donor AdoMet for the transmethylation of proteins, nucleic acids and other molecules, with far-reaching regulatory roles. Along the route it also yields homocysteine whose own metabolism is at the crossroads of several pathways. The salvage arm of the methionine cycle leads to the decarboxylated form of AdoMet, S-adenosylmethioninamine, which is used in the biosynthesis of spermidine and spermine polyamines. Click here to explore the details of the methionine cycle. |
Homocysteine metabolism is at the crossroads of three metabolic pathways. Two remethylation pathways regenerate methionine and therefore both its cycle and homocysteine. One route is cobalamin-dependent and requires folate (5-methylTHF) as the one-carbon (1C) donor, the other is independent of cobalamin but depends on betaine as the 1C donor. The third route involves the irreversible degradation of homocysteine to cysteine and further downstream metabolites. Click here to explore the overall aspects of homocysteine metabolism. |
| Folate cycle metabolic pathway | Polyamine metabolic pathway |
| The folate cycle and the folate-mediated one-carbon pathways are part of the folate metabolic pathways. The metabolic cycle deals with the various facets involving transport, modifications and interconversions of folates. Click here to explore this critical aspect of folate metabolism. |
The polyamine cations interact with nucleic acids, proteins and other
|
Return to pathway suite network diagram
SAM-homeostasis-related Regulatory Pathway Suite
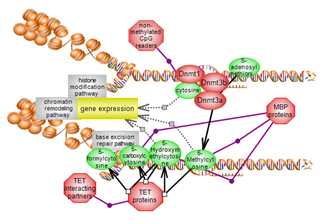
| DNA modification pathway | |
| The DNA modification pathway involves the methylation of cytosines within CpG genomic dinucleotides. Methylated and non-methylated CpGs are binding partners for numerous players. Members of the TET family of dioxygenases act upon the methylated cysteine and are also binding partners. A complex interplay exists between DNA methylation and histone modification, particularly histone lysine methylation. Click here to explore this important epigenetic pathway. |
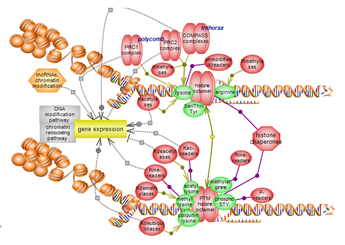
The histone modification pathway involves several residues, particularly lysines, and numerous modification types. Many classes of enzymes are involved in the various modification types while others act to remove them. Several domain types participate in the recognition of modified residues with unmodified also acting as binding partners. A complex interplay exists between histone modification, particularly histone lysine methylation, and DNA methylation. Click here to explore several aspects of this very important epigenetic pathway. |
Click here to view the Methionine, Homocysteine, Folate and Related Metabolites Pathway Suite.
Click here to view the Gene Expression and Regulation Pathway Suite Network.
Return to pathway suite network diagram
|
|
||
|
|
|
|
|
|
||