RGD Strains
What is a strain?
Rats that are used in a laboratory come from different sources; most of these are derived from Rattus norvegicus. An inbred strain is one whose background can be defined and is named by nomenclature conventions. Normally, strains are. Normally, strains are named following the nomenclature guidelines. The official designation for a rat strain is a unique, brief symbol made up of letters or a combination of letters and numbers, and a laboratory code.
 |
What types of strains does RGD have information about?RGD currently houses information about strains of the following types: advanced intercross line chromosome altered coisogenic strain congenic strain conplastic strain consomic strain hybrid strain inbred strain mutant strain outbred strain recombinant inbred strain segregating inbred strain transchromosomal strain transgenic strain wild |
|
|
HINT:The Annotation icon will bring users to the ontology annotations report page to view all objects associated with the term. |
Click here for definitions of the rat strain types.
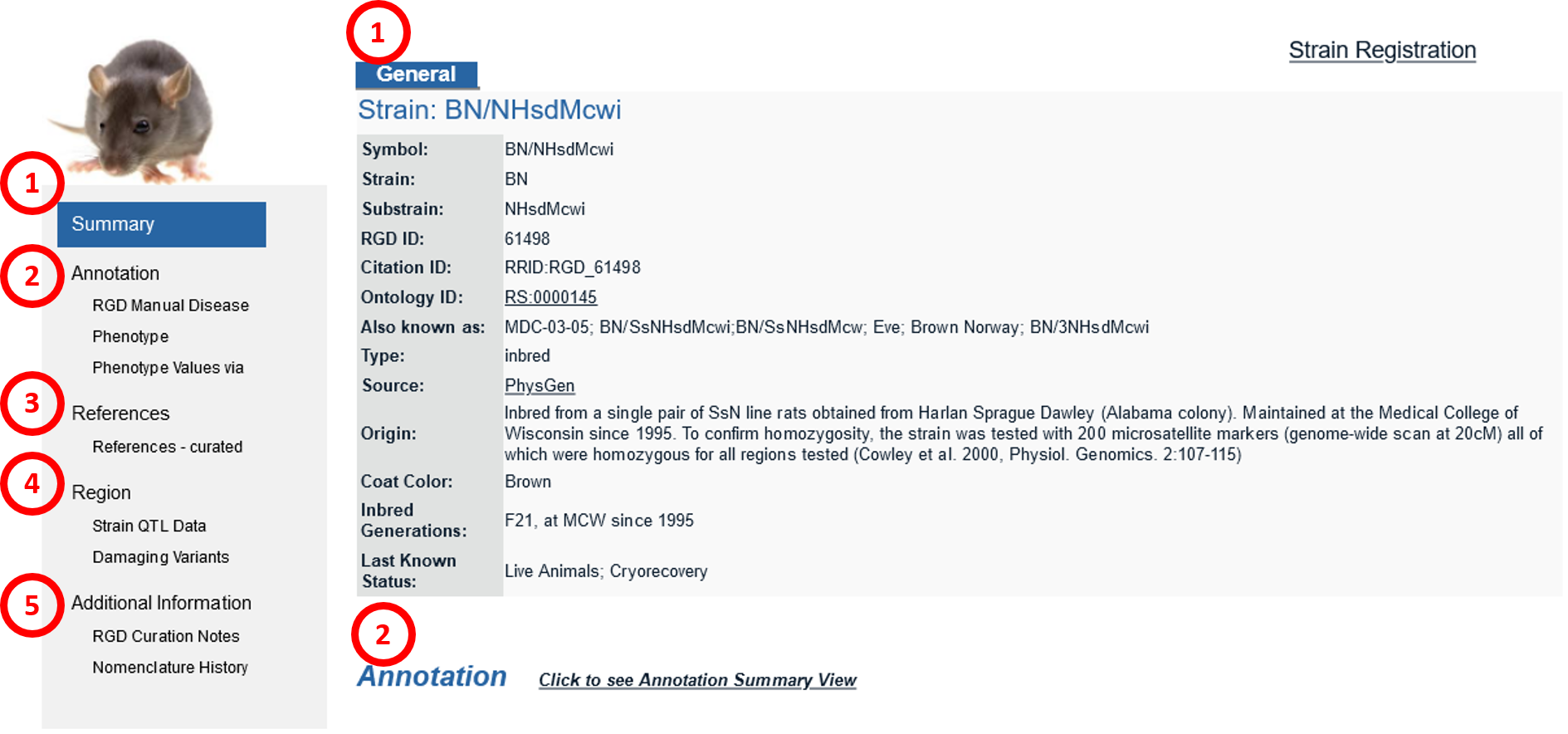
Coat Color: The coat color of this strain – e.g. Albino, brown, white. Inbred Generations: The number of generations that this strain has been inbred at the time the data was curated. Genetic status: The genotype of the strain. Last known status: The availability of the strain as last known. Position: The chromosomal positions of the genetic markers this strain carries.
2. AnnotationThis section has subsections containing ontology annotations assigned to the strain. These are organized by ontology type, which includes the Disease Ontology and the Mammalian Phenotype ontology. The phenotype values via PhenoMiner contain information about quantitative phenotype analysis in the PhenoMiner tool. The default is the “Detail View” and can be changed to the “Summary View” for easy navigation.
3. References4. RegionThis section contains Strain QTL Data and Damaging Variants analyzed from the tool Variant Visualizer. If it is a congenic strain, the Position Markers are listed in this section.
|
||||||||||||||||||||||||||||||||||