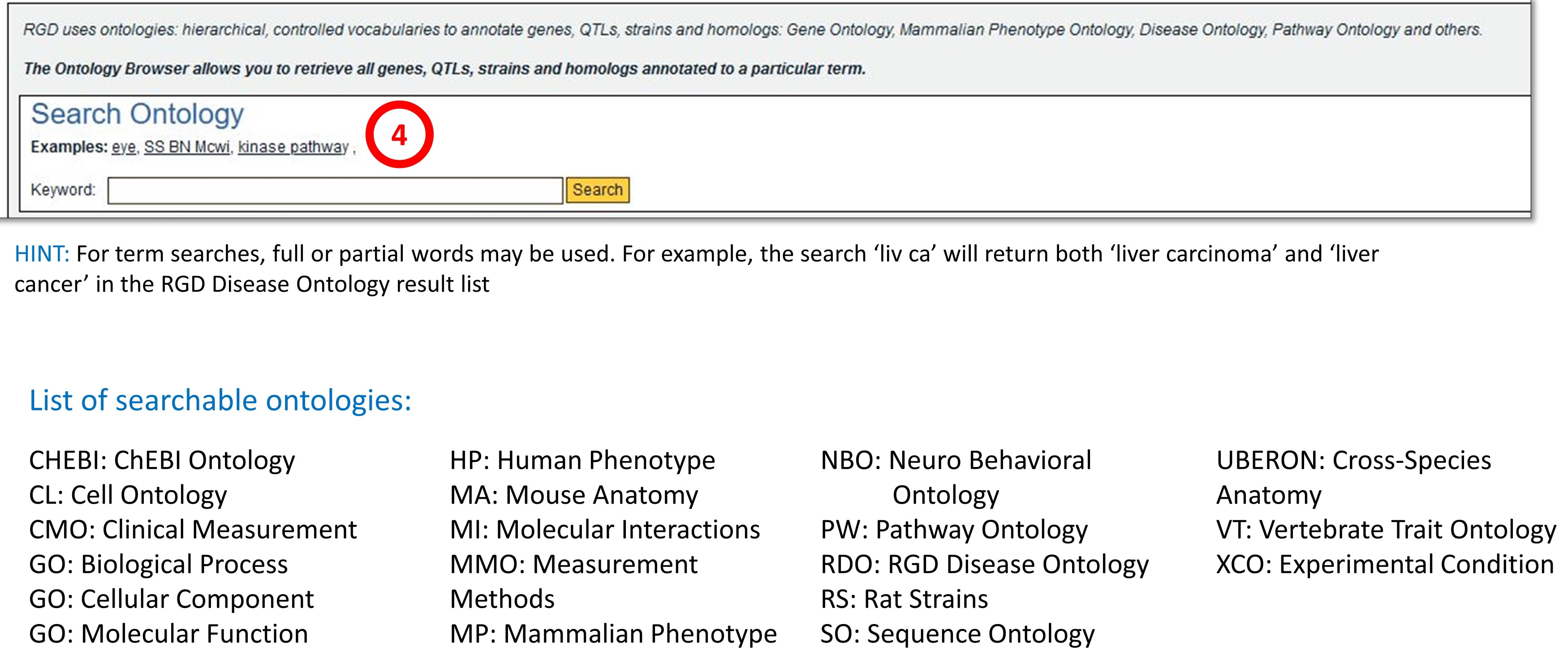
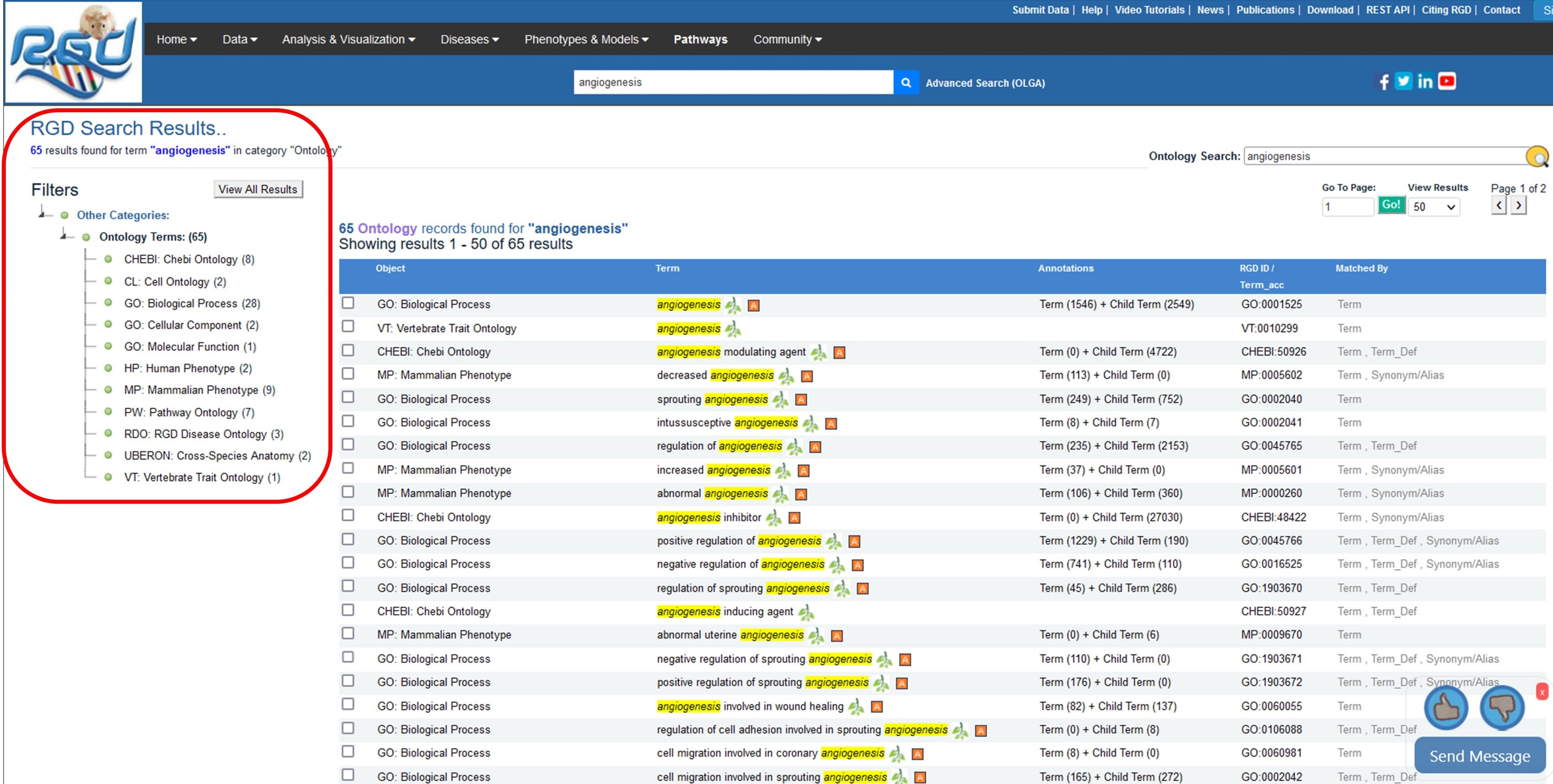
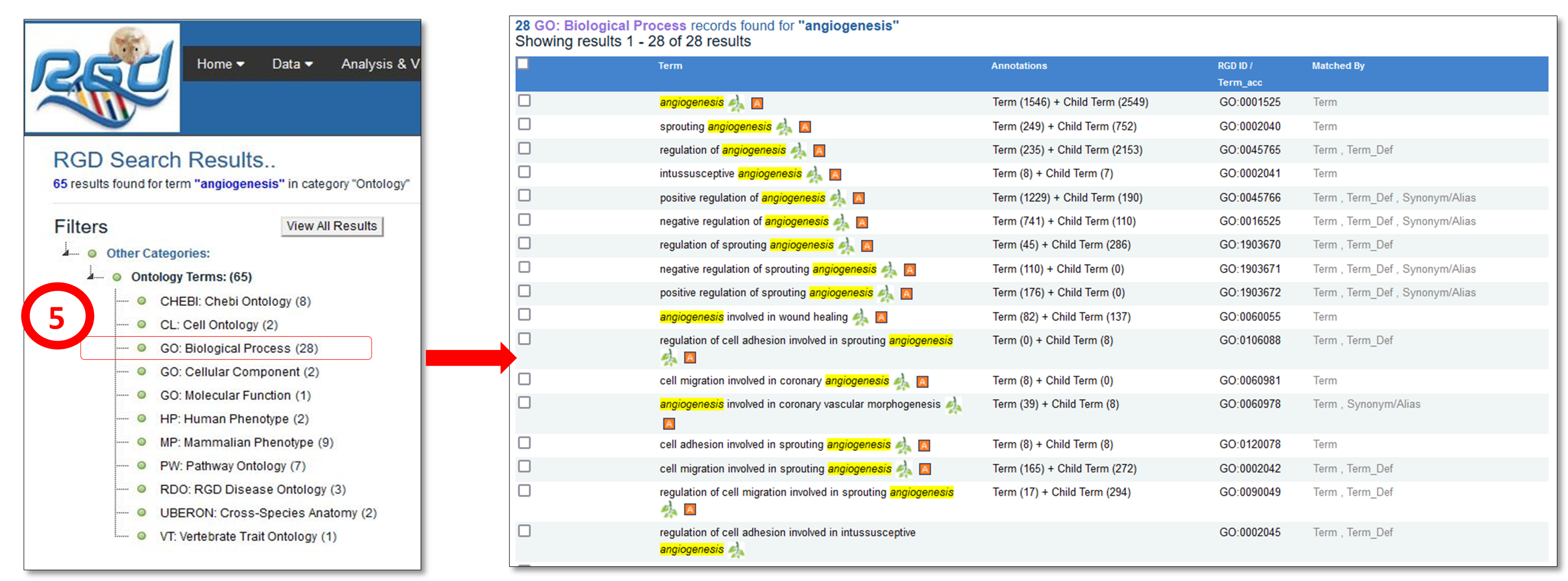
RGD uses ontologies to standardize curated data and provide that data to users in an organized manner. Over time the ontologies used at RGD have grown in number to provide more comprehensive data. The first implemented ontology at RGD was the Gene Ontology. It uses controlled vocabularies in three aspects – biological process, cellular component and molecular function, to describe genes and gene products. To increase the granularity of disease vocabularies so more specific information can be curated RGD utilizes a hybrid of Disease Ontology and RGD Disease Ontology terms. RGD has created the Pathway Ontology for gene and disease curation, and four ontologies: Clinical Measurement Ontology, Measurement Method Ontology, Experimental Condition Ontology, and Rat Strain Ontology, to curate and display quantitative phenotype data in a standardized manner. Small (not directly encoded by the genome) chemical compounds are included through the addition of the Chemical Entities of Biological Interest Ontology (ChEBI) ontology developed by the European Bioinformatics Institute and Mammalian and Human Phenotype Ontologies are also included. In collaboration with Mouse Genome Informatics at the Jackson Laboratory and the Animal QTL database at Iowa State, RGD has also created the Vertebrate Trait Ontology which includes measurable or observable characteristics in relation to morphology, physiology, or development of vertebrate organisms.
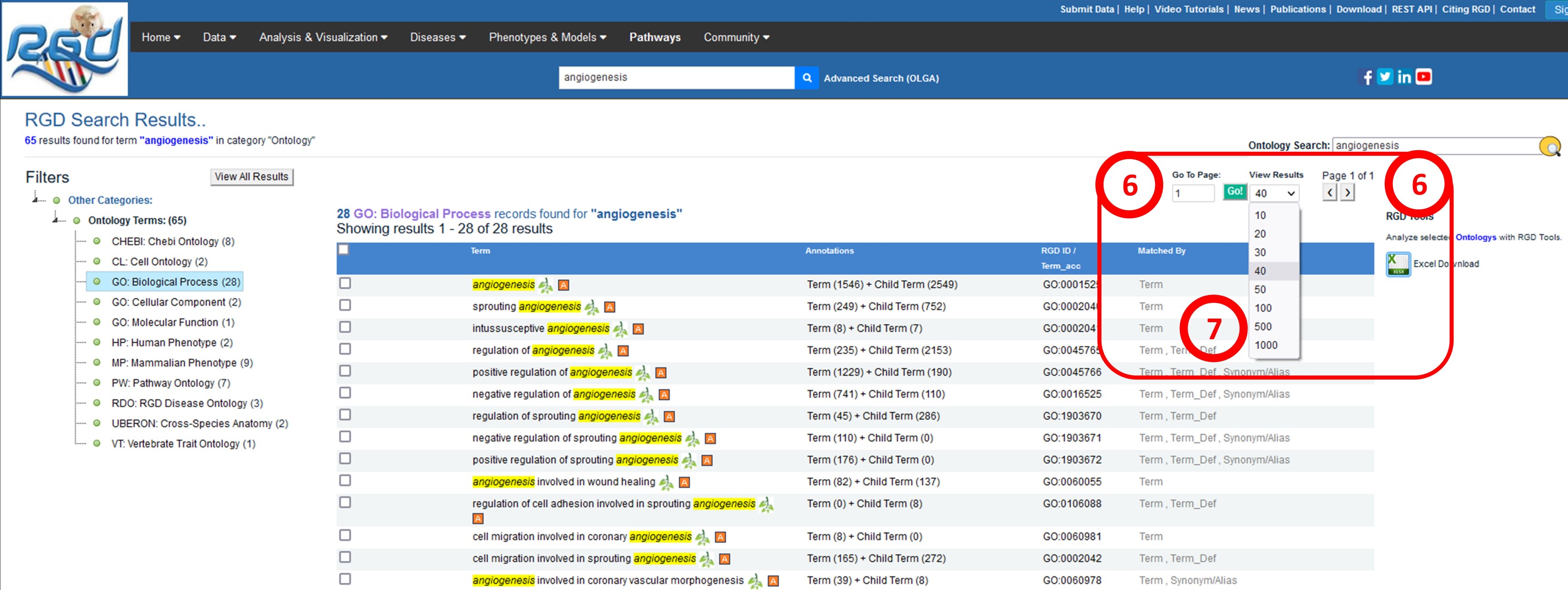
Ontology Search
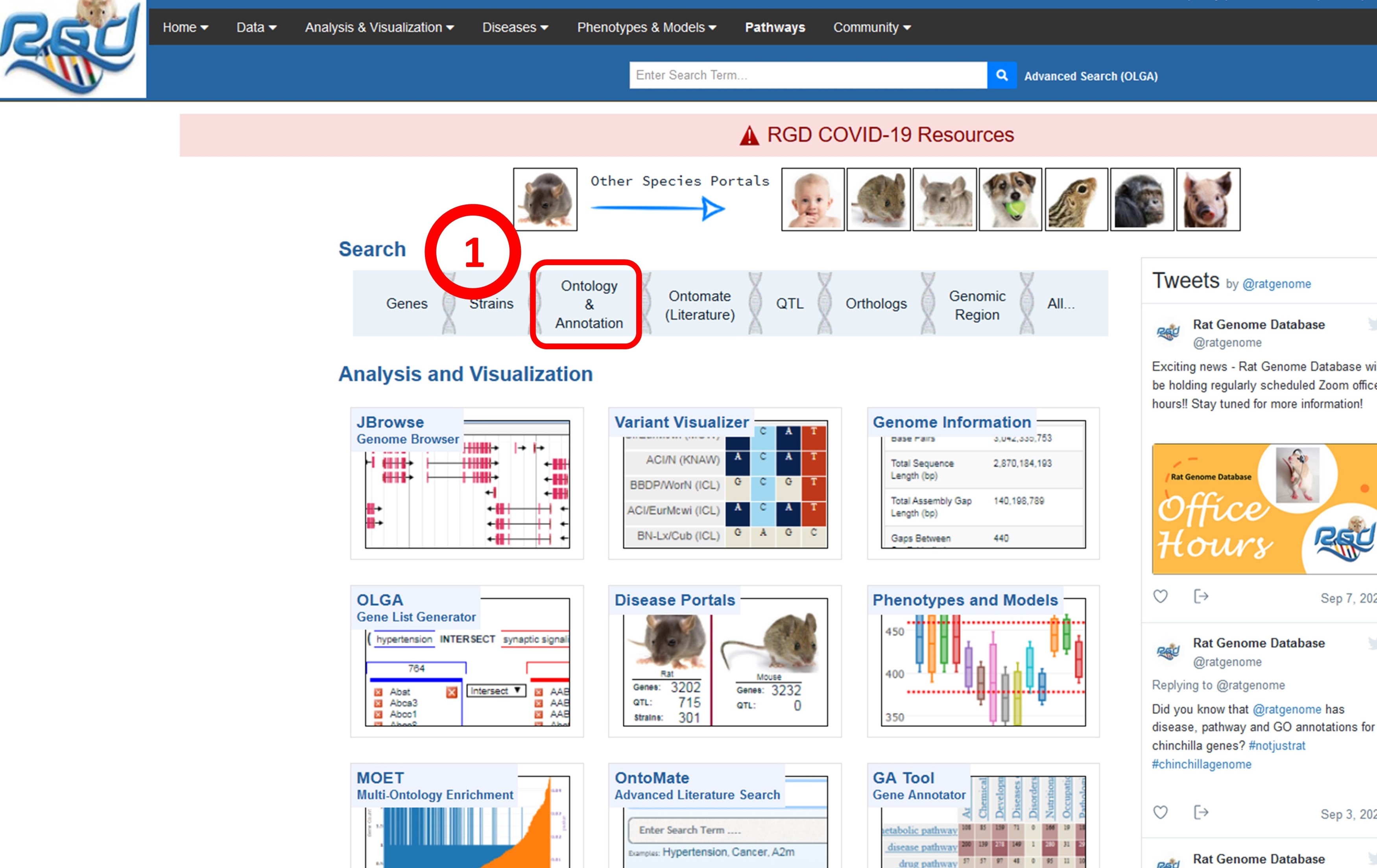 |
1. The Ontology and Annotation landing page can be directly accessed from the RGD home page.
|