Gene Report Help Page
Below is a description of each of the information sections available on a gene report page. Not all information will be available for every gene.
For information on official rat gene nomenclature click here.
Access to Gene Report Help Page
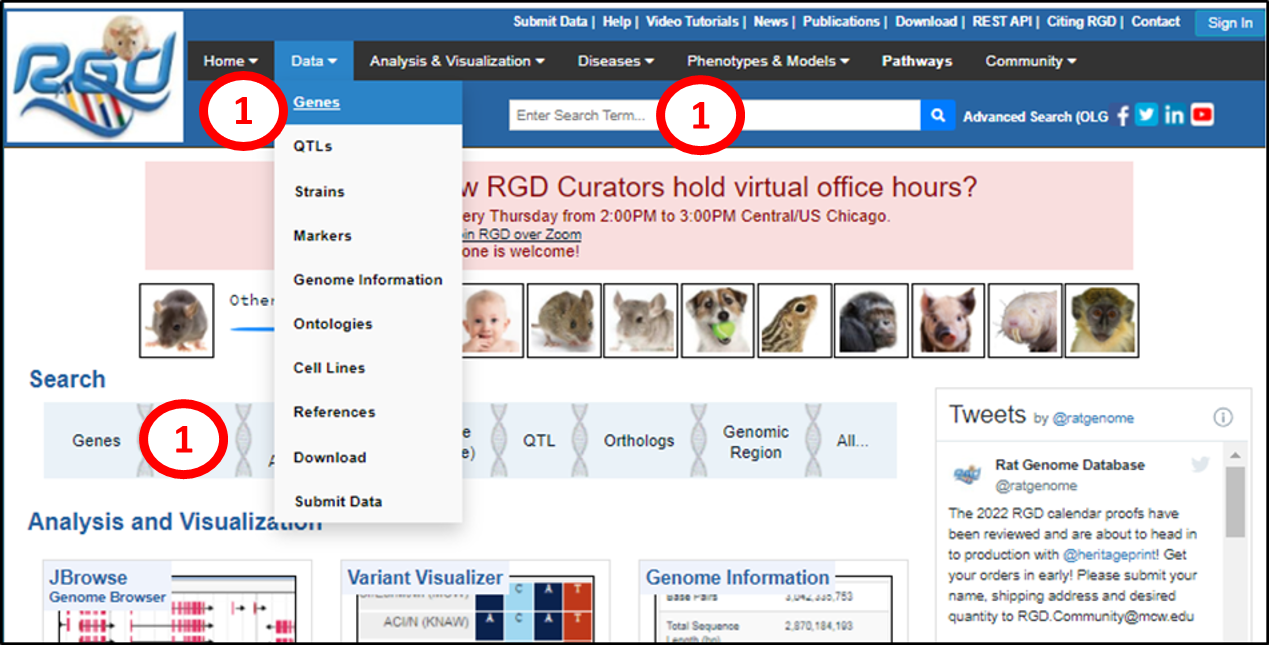 |
1. Gene report pages can be accessed from the RGD front page in several ways. One can type a case-insensitive gene symbol or name into the “Search Term” box and press enter or click the magnifying glass icon, or click on “Genes” in the “Data” pulldown in the black toolbar, or on “Genes” in the light blue Search bar.
|
 |
2. The “Search Term” method leads to an intermediate results page displaying objects in any of the ten RGD species, ontology terms or references containing the search query. Clicking on any highlighted number or phrase links to a list of matches for that category. To view the list of rat genes that match query “tnf”, click the “82” under “Rat” and see #5.
|
 |
3. The second two gene search methods described in #1 link to the Gene Search page, where one can type in gene symbols, names or ID numbers, as indicated in the examples, and press “Enter” or click “Search Genes”.
4. The search can also be limited by chromosome, start and stop base pair coordinates and species, with rat as the default.
|
 |
5. The search result for this, and from #2 above, is a results page of genes matching the search term (highlighted in yellow), with matching alleles, protein-coding regions, pseudogenes and splice variants (not shown) also accessible in the Filters at the left. Each gene is displayed with additional information explained in the blue bar at the top of the list. Multiple genes may be selected with the check boxes to the left of the gene symbols and downloaded or analyzed with RGD tools using links at the far right.
6. The search can be limited to a particular genome assembly, can be sorted by different parameters, and can be viewed by page or number of results.
7. Clicking the gene symbol links to that gene’s report page. All object report pages at RGD have a format similar to that of the gene report page.
|
The Gene Report Page Summary I (here rat Tnf)
 |
8. Gene Symbol, Name and Species and Gene Page Tabs: The official gene symbol, official gene name and species name and species picture are prominently displayed at the top of the gene page. On the left side is a floating outline of the report page sections and subsections, with the one currently being viewed highlighted in dark blue. These can be accessed directly by clicking the section title or by scrolling down. The “General” tab is selected by default and contains most of the information in RGD about the gene. For genes with microarray IDs a second tab appears next to the “General” tab. The “Array IDs” tab contains lists of microarray probe IDs associated with the gene. Currently RGD imports probe to gene associations from Affymetrix and Ensembl. Probe lists are separated according to the source.
9. Description: An automated, current synopsis of specific gene information, based on annotations of the function, processes, pathways, cellular location, diseases, phenotypes and chemical interactions associated with the gene/gene product. The information may be either experimentally derived or inferred from rat, mouse or human orthologs.
10. Type: A one-word description to designate the gene as protein-coding, pseudogene, etc. If the gene report represents a splice variant or an allele, a link to the parent gene page will be included.
RefSeq Status: A one-word code related and linked to the NCBI RefSeq record and the GenBank sequence data that were used to provide the record.
Also known as: Symbols and synonyms that are also used or have been used to refer to this gene.
|
|
The Gene Report Page Summary II
 |
11. RGD Orthologs: RGD stores data for rat, mouse, human, chinchilla, dog, bonobo, thirteen-lined ground squirrel, pig, green monkey and naked mole rat genes and for the orthologous relationships between them for comparative purposes. For rat, the “RGD Orthologs” line will contain icons representing all available orthologs in the nine other species housed at RGD. Mousing over the symbol displays the common name of the species and clicking on it links to the corresponding RGD gene report page.
12. Alliance Genes: Icon links to member species orthologous gene pages at the Alliance for Genomic Resources are provided.
13. More Info: Clicking on the gray box expands a section with more information about RGD and Alliance orthologs.
14. Is Marker For: Strains and QTLs where the gene or a mutant version is a marker are listed with links to their report pages.
Allele / Splice (not shown): Links to mutant allele and splice variant report pages are provided.
Genetic Models (not shown): Shown are rat mutant model strains that are linked to their report pages.
15. Latest Assembly: The latest genome assembly is indicated, which for rat is mRatBN7.2.
16. Position: The gene chromosomal coordinates in each genome assembly is given, with the current assembly highlighted and linked to an NCBI information page. Assembly source and genome browser links are also provided.
17. JBrowse: & Model: a link is provided to JBrowse showing the chromosomal region of the gene. This can also be accessed by clicking “Full-screen view” in the sample JBrowse window.
|
 |
18. Annotation section: annotations made using six ontologies are displayed in ‘Annotation Detail View” by default. The summary view is displayed by clicking the link. The annotation section titles are listed in the floating outline at the left and can be accessed by clicking those or scrolling down.
19. For each section 20 annotations are displayed by default (unless fewer are present), with the total number of annotations indicated. These can be scrolled through with the blue arrows, and the number displayed can be changed in the pulldown. Any word, phrase or number can be entered in the “Search table” box at the right and all annotations in the section containing what is entered will be displayed.
20. The annotations are in order alphabetically by term, and the list can be sorted alphabetically or numerically by any of the categories using the black arrows. Each annotation line consists of a term, linked to its annotation report page, a qualifier (list?), an evidence code (list), a “With” field, usually containing the RGDID of the species annotated, a reference, linked to the manuscript used to make the annotation or a link to an information page on the pipeline used to import the annotation, a “Notes” field, a database “Source” field and an “Original Reference(s)” field containing a link to the annotation’s manuscript for imported annotations. If a link is used to navigate away from the report page, upon returning to it, the report page will be configured the way it was when it was left.
|
 |
21. References section: this is composed of two subsections, “curated”, references used to make annotations, and “Additional”, uncurated references associated with the gene. All are linked to their PubMed abstracts and manuscripts.
22. Genomics section: this is comprised of five subsections, “Candidate Gene Status”, displayed when the gene is a candidate gene for one or more QTLs, with links to their report pages, “Comparative Map Data”, displaying genome assembly information similar to the “Position” section above (#16) for all of the species housed at RGD, “Position Markers”, showing genome information for markers associated with the gene, linked to their report pages, “QTLs in Region (mRatBN7.2)”, a downloadable list of QTLs that overlap the gene, linked to their report pages, presenting information about them in a section with functionality similar to the annotation sections and “miRNA Target Status”, which provides downloadable information on miRNAs predicted to target the gene.
|
 |
23. Expression section (information displayed is for demonstration purposes, not Tnf data): this shows RNA-SEQ expression data at four pre-defined levels in several biological systems. The numbers in each expression level/system category are linked to a list of that subset of data. The entire list can be viewed by clicking “View RNA-SEQ Expression Data”. All lists are downloadable.
24. Sequence section: this contains two to five subsections, “Nucleotide Sequences”, presenting available transcript and genomic sequences for the gene, viewable in different linked formats plus links to gene-associated NCBI GEO profiles for each sequence and reference sequence information, a similar “Protein Sequences” subsection, “Transcriptome” (rat gene report pages), providing links to eQTL, Weighted Gene Co-expression Network Analysis and tissue/strain expression data at Phenogen, “Protein Domains” (not shown), providing links to other genes containing domains found in the protein, “Promoters” (human and mouse gene report pages), listing positional and other linked information about the gene’s promoter(s), “Damaging Variants” (dog gene report pages) and “Clinical Variants” (human gene report pages), providing a searchable (human) list of mutations, linked to their report pages (human) with positional and descriptive information with links.
|
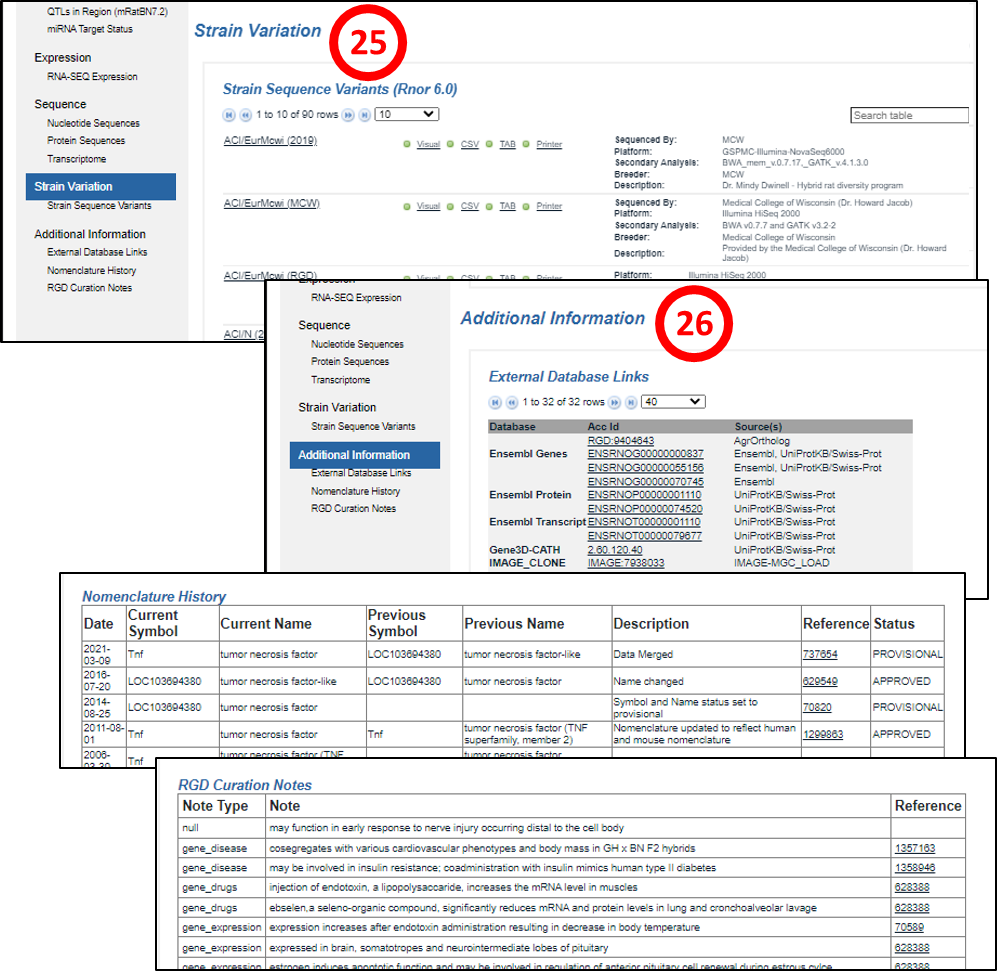 |
25. Strain Variation section (rat gene report pages): provides a list of rat strains, linked to their report pages, carrying mutations in the gene plus sequencing, analysis and source information. The mutation data are downloadable in several formats.
26. Additional Information section: this is composed of one to three subsections, “External Database Links”, listing a variety of databases with links that house different kinds of data on the gene, “Nomenclature History” (rat and human gene report pages), providing the gene name status and any previous names and “RGD Curation Notes” (rat gene report pages), legacy curator-provided descriptive information on the gene.
|
[return to top]
|



