About Ontologies
Ontology Help |
|
| The Rat Genome Database is a rich and growing repository of biomedical research data using Rattus as the model organism. To extend the usability of RGD, approaches as to how the information in the database is grouped, cataloged and presented have been and are being developed. One approach for knowledge representation is the implementation of ontologies that classify related concepts within hierarchies. At present we use sixteen ontologies as a means to present and view data in the RGD website. | |
| What is an ontology? | |
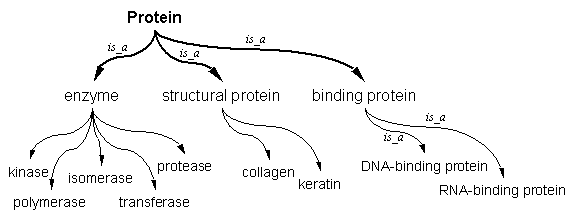
| Ontologies, as used in a biomedical context, are controlled vocabularies in which a set of related concepts and ideas that are relevant to a field are organized in a hierarchical fashion, similar to an outline. The more general concepts are placed in the higher levels and more specific concepts are assigned to lower levels. In a simple ontology of concepts relating to proteins, we might find the following terms: protein, enzyme, structural proteins, kinase, polymerase, binding protein, isomerase, collagen, transferase, keratin, DNA-binding protein, RNA-binding protein, protease. In the ontology, they might be organized as follows: Protein would be at the top of the hierarchy being the most general concept. Directly under that would be enzyme, structural protein, and binding protein. Under enzyme then would be the concepts kinase, polymerase, isomerase, transferase, protease. Similarly, collagen and keratin would fall under structural protein, etc.
The above organization illustrates an important feature of ontologies: concepts have relationships between them. In the above case enzyme is a parent concept and kinase is one of its children. Kinase is a more specific type of enzyme and the type of relationship they have is called an is_a relationship. Other relationship types can be defined for ontologies, but the other more common type is called the part_of relationship. One illustration of the latter would be the stomach is part_of the digestive system in an ontology of anatomical terms. Because of the complexity of biomedical data, the simple outline-like hierarchy used in the example is usually insufficient to capture biological knowledge, so the rules in ontology creation allow for concepts to have multiple parents along with multiple children (see below figure). To expand the original illustration, protein can have a parent, biomolecule. Biomolecule in turn can have a child catalytic biomolecule. Catalytic biomolecule in turn can be a parent of enzyme along with protein. This is allowable as long as the concepts increase in specificity as they go lower in the hierarchy and that no concept is the parent of its own ancestor. That constraint confines these ontologies to what is called a directed acyclic graph (DAG) structure.
As a hierarchy of related concepts, ontologies provide an ideal framework onto which data and information can be organized. Typically, specific examples, or instances, are linked, or annotated, to the concepts. An ontology with annotations is called a knowledge base. RGD uses ontologies to provide new avenues by which the user can find, and focus on, its objects’ information. Currently the gene ontology and fifteen other ontologies are implemented to give another contextual framework for RGD gene, quantitative trait locus and strain objects as well as keywords for RGD’s general search. [ back to top ] |
|