NON-SMALL CELL LUNG CANCER PATHWAY (PW:0000705)
Description
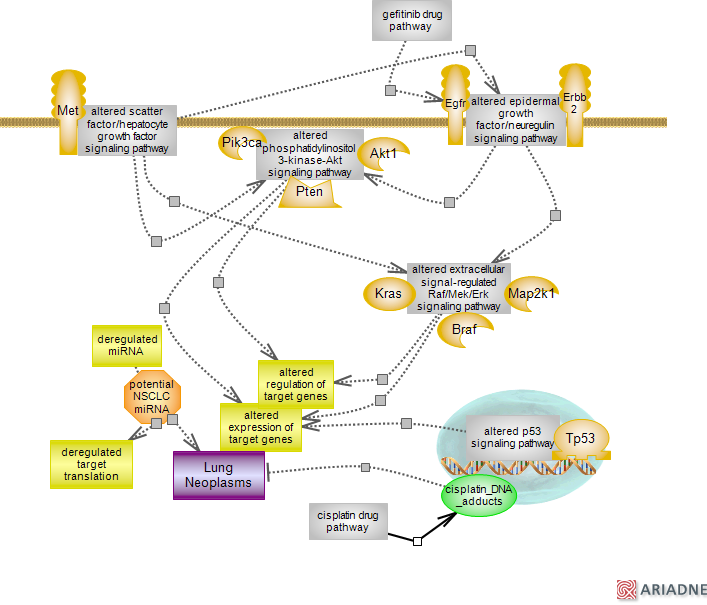
Lung cancer, a major cause of cancer mortality, has been assigned to two types: small-cell (SCLC) and non-small cell lung cancer (NSCLC). The first, an extremely aggressive form of� cancer accounts for ~20% of cases while the second accounts for the remaining ~80%. NSCLC itself is comprised of three subtypes: squamous- and large-cell carcinoma and adenocarcinoma. Of the three, the most prevalent type is adenocarcinoma. Well documented contributors to NSCLC are mutations in the epidermal growth factor receptor Egfr of the epidermal growth factor and in Kras of the Raf/Mek/Erk signaling pathway, respectively. Mutations in Egfr remove an autoinhibitory effect, lead to increased phosphorylation and to ligand-independent dimerization of the receptor. Mutations that render the receptor constitutively active also confer higher affinity for drugs such as gefitinib or erlotinib. Known as tyrosine kinase inhibitors (TKI), the drugs compete with ATP for binding to the tyrosine kinase pocket of Egfr (other drugs include monoclonal antibodies that target the ligand binding domain and compete with the endogenous ligands). The efficiency of TKI is however lost within one year of treatment due to acquired resistance. Secondary mutations in Egfr, such as the T790M substitution reported in ~50% of cases and amplification of Met receptor observed in ~20% of cases are thought to be contributing factors. T790M mutation lies within the ATP binding site and is thought to interfere with drug binding; Met amplification appears to involve activation of Erbb3. Note that Egfr and Met mediated signaling activate PI3K-Akt pathway; within the Egf context, Erbb3 is the main effector while under normal conditions Met does not engage Egfr members. Mutations in Kras lead to irreversible GTP binding resulting in its constitutive activation. Other culprits include Erbb2, Pik3ca, Akt1, Braf and Map2k1. Like Egfr, Erbb2 is a receptor for epidermal growth whose NSCLC associated mutations give rise to a constitutively active receptor. Mutations in Pik3ca, the catalytic subunit of class IA lipid kinase PI3K, and in the Akt1member of the AKT kinase family lead to gain of function. Mutant Pik3ca leads to Akt activation in the absence of growth factor stimulation, the activity of mutant Akt1 is independent of upstream PI3K function. Mutations in Braf lead to increased kinase activity while Map2k1, downstream of Braf shows gain of function in vitro in these members of the Raf/Mek/Erk cascade. At the same time, mutations in known tumor suppressors such as Tp53 or Pten lead to loss of activity thus abrogating their tumor suppressor capacity. In addition to the TKIs mentioned, other drugs are being developed that target these and other mutants associated with NSCLC and mentioned in the text. NSCLC is one of the tumor types for which platinum (Pt)-containing drugs such as cisplatin are used as chemotherapy agents.
To see the ontology report for annotations GViewer and download, click here To see the Disease Ontology report for genes that in some fashion have been associated with NSCLC or suspected to confer susceptibility,
click here To link to the Cancer Disease Portal,
click here To link to the Respiratory Disease Portal,
click here [click to see the associated KEGG map -
map05223 ]
...(less)
Pathway Diagram:
Genes in Pathway:
G
Akt1 AKT serine/threonine kinase 1
ISO IEA KEGG PMID:21277552 rno:05223, RGD:5133243
NCBI chr 6:131,713,716...131,735,319
G
Akt2 AKT serine/threonine kinase 2
IEA KEGG rno:05223
NCBI chr 1:82,877,228...82,933,828
G
Akt3 AKT serine/threonine kinase 3
IEA KEGG rno:05223
NCBI chr13:88,943,708...89,225,831
G
Bad BCL2-associated agonist of cell death
IEA KEGG rno:05223
NCBI chr 1:204,133,502...204,142,829
G
Braf B-Raf proto-oncogene, serine/threonine kinase
ISO IEA KEGG PMID:20951313 PMID:21277552 rno:05223, RGD:5133242 , RGD:5133243
NCBI chr 4:68,375,484...68,510,652
G
Casp9 caspase 9
IEA KEGG rno:05223
NCBI chr 5:154,108,872...154,126,628
G
Ccnd1 cyclin D1
IEA KEGG rno:05223
NCBI chr 1:200,089,002...200,098,524
G
Cdk4 cyclin-dependent kinase 4
IEA KEGG rno:05223
NCBI chr 7:62,886,124...62,889,562
G
Cdk6 cyclin-dependent kinase 6
IEA KEGG rno:05223
NCBI chr 4:30,637,650...30,829,688
G
Cdkn2a cyclin-dependent kinase inhibitor 2A
IEA KEGG rno:05223
NCBI chr 5:103,984,949...103,992,143
G
E2f1 E2F transcription factor 1
IEA KEGG rno:05223
NCBI chr 3:143,064,535...143,075,362
G
E2f3 E2F transcription factor 3
IEA KEGG rno:05223
NCBI chr17:34,601,756...34,679,139
G
Egf epidermal growth factor
IEA KEGG rno:05223
NCBI chr 2:218,219,408...218,302,359
G
Egfr epidermal growth factor receptor
ISO IEA KEGG PMID:20519133 PMID:20951313 rno:05223, RGD:5133241 , RGD:5133242
NCBI chr14:91,176,979...91,349,722
G
Erbb2 erb-b2 receptor tyrosine kinase 2
ISO IEA KEGG PMID:21277552 rno:05223, RGD:5133243
NCBI chr10:83,411,197...83,435,078
G
Fhit fragile histidine triad diadenosine triphosphatase
IEA KEGG rno:05223
NCBI chr15:13,935,029...15,442,620
G
Foxo3 forkhead box O3
IEA KEGG rno:05223
NCBI chr20:45,669,708...45,764,606
G
Grb2 growth factor receptor bound protein 2
IEA KEGG rno:05223
NCBI chr10:100,881,404...100,949,193
G
Hras HRas proto-oncogene, GTPase
IEA KEGG rno:05223
NCBI chr 1:196,290,127...196,299,823
G
Kras KRAS proto-oncogene, GTPase
ISO IEA KEGG PMID:20951313 rno:05223, RGD:5133242
NCBI chr 4:178,185,418...178,218,484
G
Map2k1 mitogen activated protein kinase kinase 1
ISO IEA KEGG PMID:20951313 PMID:21277552 rno:05223, RGD:5133242 , RGD:5133243
NCBI chr 8:64,683,449...64,754,900
G
Map2k2 mitogen activated protein kinase kinase 2
IEA KEGG rno:05223
NCBI chr 7:8,590,729...8,610,279
G
Mapk1 mitogen activated protein kinase 1
IEA KEGG rno:05223
NCBI chr11:83,957,813...84,023,629
G
Mapk3 mitogen activated protein kinase 3
IEA KEGG rno:05223
NCBI chr 1:181,366,646...181,372,863
G
Met MET proto-oncogene, receptor tyrosine kinase
ISO RGD
PMID:20519133 PMID:20951313 PMID:21277552 RGD:5133241 , RGD:5133242 , RGD:5133243
NCBI chr 4:45,790,456...45,898,139
G
Nras NRAS proto-oncogene, GTPase
IEA KEGG rno:05223
NCBI chr 2:190,582,885...190,593,509
G
Pdpk1 3-phosphoinositide dependent protein kinase-1
IEA KEGG rno:05223
NCBI chr10:13,105,435...13,182,664
G
Pik3ca phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha
ISO IEA KEGG PMID:20951313 PMID:21277552 rno:05223, RGD:5133242 , RGD:5133243
NCBI chr 2:115,175,275...115,249,034
G
Pik3cb phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta
IEA KEGG rno:05223
NCBI chr 8:99,594,600...99,699,772
G
Pik3cd phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta
IEA KEGG rno:05223
NCBI chr 5:160,094,952...160,142,962
G
Pik3r1 phosphoinositide-3-kinase regulatory subunit 1
IEA KEGG rno:05223
NCBI chr 2:32,878,942...32,963,668
G
Pik3r2 phosphoinositide-3-kinase regulatory subunit 2
IEA KEGG rno:05223
NCBI chr16:18,665,517...18,674,067
G
Pik3r3 phosphoinositide-3-kinase regulatory subunit 3
IEA KEGG rno:05223
NCBI chr 5:129,700,925...129,772,591
G
Pik3r5 phosphoinositide-3-kinase, regulatory subunit 5
IEA KEGG rno:05223
NCBI chr10:53,132,585...53,200,663
G
Plcg1 phospholipase C, gamma 1
IEA KEGG rno:05223
NCBI chr 3:149,385,587...149,416,330
G
Plcg2 phospholipase C, gamma 2
IEA KEGG rno:05223
NCBI chr19:45,547,416...45,683,930
G
Prkca protein kinase C, alpha
IEA KEGG rno:05223
NCBI chr10:92,889,390...93,288,013
G
Prkcb protein kinase C, beta
IEA KEGG rno:05223
NCBI chr 1:176,832,173...177,163,539
G
Prkcg protein kinase C, gamma
IEA KEGG rno:05223
NCBI chr 1:65,832,851...65,860,676
G
Pten phosphatase and tensin homolog
ISO RGD
PMID:20951313 RGD:5133242
NCBI chr 1:230,630,443...230,697,070
G
Raf1 Raf-1 proto-oncogene, serine/threonine kinase
IEA KEGG rno:05223
NCBI chr 4:148,679,534...148,740,265
G
Rarb retinoic acid receptor, beta
IEA KEGG rno:05223
NCBI chr15:8,700,533...9,051,288
G
Rassf1 Ras association domain family member 1
IEA KEGG rno:05223
NCBI chr 8:108,225,023...108,236,164
G
Rassf5 Ras association domain family member 5
IEA KEGG rno:05223
NCBI chr13:42,637,513...42,703,024
G
Rb1 RB transcriptional corepressor 1
IEA KEGG rno:05223
NCBI chr15:48,371,295...48,502,473
G
Rxra retinoid X receptor alpha
IEA KEGG rno:05223
NCBI chr 3:10,989,832...11,076,366
G
Rxrb retinoid X receptor beta
IEA KEGG rno:05223
NCBI chr20:4,816,813...4,823,267
G
Rxrg retinoid X receptor gamma
IEA KEGG rno:05223
NCBI chr13:79,743,430...79,785,173
G
Sos1 SOS Ras/Rac guanine nucleotide exchange factor 1
IEA KEGG rno:05223
NCBI chr 6:14,533,870...14,613,348
G
Sos2 SOS Ras/Rho guanine nucleotide exchange factor 2
IEA KEGG rno:05223
NCBI chr 6:88,042,966...88,156,140
G
Stk4 serine/threonine kinase 4
IEA KEGG rno:05223
NCBI chr 3:152,745,958...152,827,747
G
Tgfa transforming growth factor alpha
IEA KEGG rno:05223
NCBI chr 4:118,618,043...118,700,897
G
Tp53 tumor protein p53
ISO IEA KEGG PMID:20951313 rno:05223, RGD:5133242
NCBI chr10:54,300,070...54,311,525
Pathway Gene Annotations
Disease Annotations Associated with Genes in the non-small cell lung cancer pathway
Akt1 Acute Lung Injury , acute promyelocytic leukemia , adenocarcinoma , Alzheimer's disease , amphetamine abuse , amyotrophic lateral sclerosis , Animal Disease Models , atherosclerosis , autosomal recessive polycystic kidney disease , bipolar disorder , bone osteosarcoma , Brain Injuries , brain ischemia , breast adenocarcinoma , breast cancer , Breast Cancer, Familial , Breast Neoplasms , Calcification of Aortic Valve , cannabis abuse , Carcinogenesis , cardiac arrest , Cardiomegaly , cardiomyopathy , Charcot-Marie-Tooth disease axonal type 2O , chronic myeloid leukemia , colon cancer , colon carcinoma , colorectal cancer , Colorectal Neoplasms , coronary artery disease , Cowden syndrome 6 , Diabetic Cardiomyopathies , diabetic retinopathy , Endometrioid Carcinomas , Endotoxemia , epilepsy , esophageal cancer , esophagus squamous cell carcinoma , Experimental Arthritis , Experimental Colitis , Experimental Diabetes Mellitus , Experimental Mammary Neoplasms , familial adenomatous polyposis , Fibrosis , focal segmental glomerulosclerosis 5 , gastric adenocarcinoma , genetic disease , glioblastoma , head and neck squamous cell carcinoma , hepatocellular carcinoma , Hepatomegaly , Hereditary Neoplastic Syndromes , Hyperplasia , hypertension , Hypertriglyceridemia , idiopathic pulmonary fibrosis , impotence , in situ carcinoma , Inflammation , intermediate coronary syndrome , Intervertebral Disc Displacement , invasive ductal carcinoma , Kidney Neoplasms , lung adenocarcinoma , Lung Neoplasms , lung non-small cell carcinoma , lung oat cell carcinoma , lung small cell carcinoma , lung squamous cell carcinoma , Macrocephaly Mesodermal Hamartoma Spectrum , malignant astrocytoma , melanoma , Memory Disorders , Meningeal Neoplasms , meningioma , muscular atrophy , myocardial infarction , Myocardial Reperfusion Injury , Neointima , obesity , osteosarcoma , ovarian cancer , Ovarian Neoplasms , Pain , pancreatic cancer , pancreatic intraductal papillary-mucinous neoplasm , paraplegia , Parkinson's disease , pre-malignant neoplasm , prostate adenocarcinoma , prostate cancer , Prostatic Neoplasms , Proteus syndrome , pulmonary tuberculosis , renal cell carcinoma , Reperfusion Injury , Right Ventricular Hypertrophy , schizophrenia , skin melanoma , Skin Neoplasms , Spinal Cord Injuries , squamous cell carcinoma , Staphylococcal Pneumonia , steatotic liver disease , Stroke , substance-induced psychosis , T-cell non-Hodgkin lymphoma , Thyroid Neoplasms , transitional cell carcinoma , type 2 diabetes mellitus , ureteral obstruction , urinary bladder cancer , Uterine Cervical Neoplasms , Ventilator-Induced Lung Injury , vulva cancer Akt2 adenocarcinoma , Breast Neoplasms , Carpenter Syndrome 2 , colorectal cancer , Colorectal Neoplasms , congenital hypoplastic anemia , craniosynostosis , Diamond-Blackfan anemia , endometrial cancer , Experimental Mammary Neoplasms , familial partial lipodystrophy , genetic disease , hepatocellular carcinoma , high grade glioma , hypertension , hypoglycemia , hypoinsulinemic hypoglycemia with hemihypertrophy , Insulin Resistance , ischemia , lung non-small cell carcinoma , malignant astrocytoma , maple syrup urine disease , maturity-onset diabetes of the young type 1 , Myocardial Reperfusion Injury , Neonatal Hypoglycemia, Simulating Foetopathia Diabetica , Neoplasm Metastasis , obesity , ovarian carcinoma , Ovarian Neoplasms , pancreatic cancer , polycystic ovary syndrome , prostate adenocarcinoma , prostate cancer , prostate carcinoma , Prostatic Neoplasms , Thyroid Neoplasms , type 2 diabetes mellitus Akt3 Bardet-Biedl syndrome , Bardet-Biedl syndrome 16 , capillary hemangioma , colon cancer , complex cortical dysplasia with other brain malformations , COVID-19 , developmental and epileptic encephalopathy 54 , Developmental Disabilities , gastrointestinal stromal tumor , genetic disease , glioblastoma , lung non-small cell carcinoma , Macrocephaly , malignant astrocytoma , megacolon , Megalencephaly - Cutis Marmorata Telangiectatica Congenita , Megalencephaly-Polymicrogyria-Polydactyly-Hydrocephalus Syndrome , Megalencephaly-Polymicrogyria-Polydactyly-Hydrocephalus Syndrome 2 , Neurodevelopmental Disorders , pancreatic adenocarcinoma , parathyroid carcinoma , polymicrogyria , prostate adenocarcinoma , prostate cancer , renal cell carcinoma , Senior-Loken syndrome , Senior-Loken Syndrome 7 Bad acute kidney failure , Alzheimer's disease , amyotrophic lateral sclerosis , Brain Injuries , brain ischemia , breast cancer , Burns , Cardiomegaly , Chemical and Drug Induced Liver Injury , Chronic Pancreatitis , colon cancer , Diabetic Cardiomyopathies , Diabetic Cystopathy , diabetic neuropathy , endometrial cancer , Experimental Diabetes Mellitus , Experimental Mammary Neoplasms , gastric ulcer , genetic disease , glaucoma , Hemorrhagic Shock , high grade glioma , hypertension , hypothyroidism , impotence , Insulin Resistance , intellectual disability , intermittent claudication , leukemia , leukocyte adhesion deficiency 3 , Liver Reperfusion Injury , lung non-small cell carcinoma , macular degeneration , middle cerebral artery infarction , Myocardial Reperfusion Injury , obesity , osteoarthritis , pancreatic cancer , Parkinsonism , prostate carcinoma , prostatic hypertrophy , Prostatic Neoplasms , pulmonary fibrosis , renal cell carcinoma , Reperfusion Injury , Retina Reperfusion Injury , Sepsis , Small-For-Size Syndrome , Spinal Cord Reperfusion Injury , transient cerebral ischemia , traumatic brain injury , type 2 diabetes mellitus , visual epilepsy Braf adrenocortical carcinoma , ameloblastoma , Aortic Aneurysm, Giant Congenital , arteriovenous malformations of the brain , ataxia telangiectasia , B-Cell Chronic Lymphocytic Leukemia , bile duct adenoma , Brain Neoplasms , Breast Neoplasms , cancer , Carcinogenesis , cardiofaciocutaneous syndrome , cardiofaciocutaneous syndrome 1 , cardiomyopathy , cholangiocarcinoma , chronic lymphocytic leukemia , chronic myelogenous leukemia, BCR-ABL1 positive , chronic myeloid leukemia , colon carcinoma , Colonic Neoplasms , colorectal adenocarcinoma , colorectal cancer , Colorectal Neoplasms , Costello syndrome , craniopharyngioma , Dandy-Walker syndrome , Developmental Disabilities , diffuse large B-cell lymphoma , dilated cardiomyopathy , disease of cellular proliferation , dysembryoplastic neuroepithelial tumor , dysplastic nevus syndrome , endometrial carcinoma , Experimental Melanoma , Experimental Neoplasms , familial melanoma , Follicular Thyroid Cancer , gallbladder cancer , ganglioglioma , gastric adenocarcinoma , gastrointestinal stromal tumor , genetic disease , germinoma , glioblastoma , head and neck squamous cell carcinoma , hepatocellular carcinoma , high grade glioma , hypertrophic cardiomyopathy , hypertrophic cardiomyopathy 4 , intellectual disability , intrahepatic cholangiocarcinoma , Kidney Neoplasms , Langerhans-cell histiocytosis , Liver Metastasis , Liver Neoplasms , lung adenocarcinoma , lung cancer , lung carcinoma , Lung Neoplasms , lung non-small cell carcinoma , lung squamous cell carcinoma , lymphangioma , Lymphatic Metastasis , malignant astrocytoma , Mandibular Neoplasms , melanoma , Microsatellite Instability , multiple myeloma , myoepithelioma , Neoplasm Metastasis , nephroblastoma , neuroendocrine tumor , Nevus , newborn respiratory distress syndrome , non-Hodgkin lymphoma , Nonseminomatous Germ Cell Tumor , Noonan syndrome , Noonan syndrome 1 , Noonan syndrome 7 , Noonan syndrome with multiple lentigines , Noonan syndrome with multiple lentigines 3 , Ovarian Neoplasms , ovarian serous cystadenocarcinoma , pancreatic acinar cell adenocarcinoma , pancreatic cancer , papillary renal cell carcinoma , Paraproteinemias , PHACE Association , plasma cell neoplasm , pleomorphic xanthoastrocytoma , Premature Birth , primary cutaneous T-cell non-Hodgkin lymphoma , prostate adenocarcinoma , Prostate Cancer, Hereditary, 1 , Prostatic Neoplasms , pulmonary valve stenosis , RASopathy , Sezary's disease , skin melanoma , Sporadic Papillary Renal Cell Carcinoma , tethered spinal cord syndrome , thyroid gland papillary carcinoma , Thyroid Neoplasms , transitional cell carcinoma , type 2 diabetes mellitus , uterine cancer , Vascular Malformations , ventricular septal defect Casp9 Acute Liver Failure , Alzheimer's disease , amyotrophic lateral sclerosis , bacterial infectious disease , Brain Hypoxia-Ischemia , brain ischemia , breast cancer , bronchopulmonary dysplasia , cataract , Cerebral Hemorrhage , chromosome 1p36 deletion syndrome , colorectal cancer , Diabetes Complications , Diabetic Cardiomyopathies , Diabetic Nephropathies , diabetic retinopathy , dilated cardiomyopathy , Edema , endometritis , Endotoxemia , Experimental Autoimmune Encephalomyelitis , Experimental Autoimmune Myocarditis , Experimental Diabetes Mellitus , Experimental Mammary Neoplasms , Experimental Radiation Injuries , focal segmental glomerulosclerosis , genetic disease , heart disease , hepatocellular carcinoma , Hereditary Pancreatitis , Huntington's disease , Hypercholesterolemia , hypertension , Intestinal Reperfusion Injury , kidney disease , liver cirrhosis , Liver Reperfusion Injury , lung non-small cell carcinoma , Lymphatic Metastasis , middle cerebral artery infarction , Myocardial Reperfusion Injury , nervous system disease , NSAID-Enteropathy , osteoarthritis , pancreatic cancer , Parkinson's disease , Parkinsonism , peripheral nervous system disease , post-traumatic stress disorder , pre-malignant neoplasm , Prostatic Neoplasms , Reperfusion Injury , retinal detachment , scrapie , Sepsis , severe acute respiratory syndrome , Spinal Cord Injuries , stomach cancer , Subarachnoid Hemorrhage , Testis Reperfusion Injury , transient cerebral ischemia , type 1 diabetes mellitus , type 2 diabetes mellitus , ureteral obstruction , urinary bladder cancer , varicocele Ccnd1 acute lymphoblastic leukemia , acute myeloid leukemia , adenocarcinoma , adenoid cystic carcinoma , Aicardi-Goutieres Syndrome 3 , angiosarcoma , Animal Mammary Neoplasms , atherosclerosis , B-lymphoblastic leukemia/lymphoma , Bowen's Disease , Brain Neoplasms , Breast Neoplasms , bronchiolo-alveolar adenocarcinoma , Burns , carcinoma , Cecal Neoplasms , cervix uteri carcinoma in situ , Chronic Hepatitis C , colon adenoma , Colonic Neoplasms , colorectal cancer , Colorectal Neoplasms , Disease Progression , ductal carcinoma in situ , Endometrial Neoplasms , Esophageal Neoplasms , esophagus squamous cell carcinoma , Experimental Liver Neoplasms , Experimental Mammary Neoplasms , Experimental Neoplasms , genetic disease , Genomic Instability , head and neck squamous cell carcinoma , hepatocellular carcinoma , high grade glioma , hyperparathyroidism , Hyperplasia , impotence , intellectual disability , Intestinal Polyps , invasive lobular carcinoma , Kidney Neoplasms , leukemia , liver cancer , Liver Neoplasms , lung adenocarcinoma , Lung Neoplasms , lung non-small cell carcinoma , lung small cell carcinoma , lung squamous cell carcinoma , malignant astrocytoma , mantle cell lymphoma , medulloblastoma , multiple myeloma , Myocardial Ischemia , Nasopharyngeal Neoplasms , Neoplasm Metastasis , Neoplastic Cell Transformation , non-Hodgkin lymphoma , ovarian carcinoma , pancreatic adenocarcinoma , pancreatic cancer , pancreatic ductal adenocarcinoma , pancreatic ductal carcinoma , Pancreatic Intraepithelial Neoplasia , pancreatic solid pseudopapillary carcinoma , papilloma , penile benign neoplasm , pre-malignant neoplasm , prostate cancer , prostate carcinoma in situ , Prostatic Neoplasms , Recurrent Infections, with Encephalopathy, Hepatic Dysfunction, and Cardiovascular Malformations , renal cell carcinoma , Reperfusion Injury , Salivary Gland Neoplasms , squamous cell carcinoma , status epilepticus , Stomach Neoplasms , thyroid gland papillary carcinoma , Thyroid Neoplasms , tongue squamous cell carcinoma , transitional cell carcinoma , urinary bladder cancer , Urologic Neoplasms , Uterine Cervical Neoplasms , visual epilepsy , von Hippel-Lindau disease Cdk4 Animal Mammary Neoplasms , Brain Neoplasms , Brain Stem Neoplasms , breast cancer , breast carcinoma , carcinoma , cervix uteri carcinoma in situ , diffuse pediatric-type high-grade glioma, H3-wildtype and IDH-wildtype , Endometrial Neoplasms , Ewing sarcoma , Experimental Liver Cirrhosis , Experimental Mammary Neoplasms , Experimental Neoplasms , familial melanoma , Germ Cell and Embryonal Neoplasms , glioblastoma , head and neck squamous cell carcinoma , hepatocellular carcinoma , Hereditary Neoplastic Syndromes , high grade glioma , Hyperoxia , islet cell tumor , Leydig cell tumor , liposarcoma , lung adenocarcinoma , lung non-small cell carcinoma , lung squamous cell carcinoma , melanoma , multiple myeloma , nephroblastoma , non-functioning pancreatic endocrine tumor , obesity , ovarian cancer , Ovarian Neoplasms , pancreatic adenocarcinoma , pancreatic cancer , pancreatic ductal adenocarcinoma , prostate cancer , prostatic hypertrophy , renal cell carcinoma , skin melanoma , stomach cancer , Stomach Neoplasms , transitional cell carcinoma , Triple Negative Breast Neoplasms , type 1 diabetes mellitus , type 2 diabetes mellitus , urinary bladder cancer , Uterine Cervical Neoplasms Cdk6 acute lymphoblastic leukemia , acute myeloid leukemia , anemia , atrial fibrillation , Behcet's disease , Brain Stem Neoplasms , Chloracne , Chromosome Aberrations , Experimental Mammary Neoplasms , genetic disease , glioblastoma , head and neck squamous cell carcinoma , high grade glioma , lung adenocarcinoma , lung non-small cell carcinoma , malignant mesothelioma , medulloblastoma , non-functioning pancreatic endocrine tumor , pancreatic adenocarcinoma , pancreatic ductal carcinoma , pleomorphic xanthoastrocytoma , primary autosomal recessive microcephaly 12 , rheumatoid arthritis , Stroke , Triple Negative Breast Neoplasms Cdkn2a acute lymphoblastic leukemia , acute myeloid leukemia , adenocarcinoma , Adrenal Gland Neoplasms , adrenocortical carcinoma , B-lymphoblastic leukemia/lymphoma , bone disease , Brain Neoplasms , Brain Stem Neoplasms , breast cancer , Breast Neoplasms , Carcinogenesis , cervix uteri carcinoma in situ , cholangiocarcinoma , Chromosome Deletion , chronic myeloid leukemia , chronic obstructive pulmonary disease , Colonic Neoplasms , Colorectal Neoplasms , diffuse large B-cell lymphoma , disease of cellular proliferation , dysplastic nevus syndrome , endometrial cancer , Endometrial Intraepithelial Neoplasia , Endometrial Neoplasms , esophageal carcinoma , Esophageal Neoplasms , esophagus adenocarcinoma , esophagus squamous cell carcinoma , Experimental Arthritis , Experimental Leukemia , Experimental Mammary Neoplasms , familial melanoma , Fluoride Poisoning , gastric adenocarcinoma , genetic disease , Genetic Translocation , Germ Cell and Embryonal Neoplasms , germinoma , glaucoma , glioblastoma , granulosa cell tumor , head and neck squamous cell carcinoma , hepatocellular carcinoma , Hereditary Neoplastic Syndromes , high grade glioma , Hyperplasia , hypertension , in situ carcinoma , invasive ductal carcinoma , Kidney Neoplasms , leiomyosarcoma , leukemia , Liver Neoplasms , lung adenocarcinoma , Lung Neoplasms , lung non-small cell carcinoma , lung squamous cell carcinoma , Lymphatic Metastasis , lymphoma , Maffucci syndrome , malignant mesothelioma , melanoma , melanoma and neural system tumor syndrome , Melanoma-Pancreatic Cancer Syndrome , Mesothelioma , Micronuclei, Chromosome-Defective , Mouth Neoplasms , multiple myeloma , myelodysplastic syndrome , Neoplasm Invasiveness , Neoplasm Metastasis , nephroblastoma , Nerve Sheath Neoplasms , neurilemmoma , neuroblastoma , obesity , osteoarthritis , osteosarcoma , ovarian cancer , Ovarian Neoplasms , pancreatic adenocarcinoma , pancreatic cancer , pancreatic carcinoma , pancreatic ductal carcinoma , papillary renal cell carcinoma , papillomavirus infectious disease , peripheral nervous system disease , persistent hyperplastic primary vitreous , plasmacytoma , Poisoning , pre-malignant neoplasm , Premature Aging , primary cutaneous T-cell non-Hodgkin lymphoma , prostate adenocarcinoma , prostate cancer , Prostatic Neoplasms , renal cell carcinoma , retinoblastoma , rhabdomyosarcoma , sarcomatoid mesothelioma , schizophrenia , Sezary's disease , skin melanoma , Sporadic Papillary Renal Cell Carcinoma , squamous cell carcinoma , stomach cancer , Stomach Neoplasms , T-cell acute lymphoblastic leukemia , thymoma , Tongue Neoplasms , transient cerebral ischemia , transitional cell carcinoma , Trisomy , urinary bladder cancer , Urinary Bladder Neoplasm , Urogenital Neoplasms , uterine carcinosarcoma , Uterine Cervical Neoplasms , Vulvar Lichen Sclerosus , Vulvar Neoplasms E2f1 Alzheimer's disease , Breast Neoplasms , Chemical and Drug Induced Liver Injury , chronic myeloid leukemia , COVID-19 , dementia , disease of cellular proliferation , genetic disease , glioblastoma , hepatocellular carcinoma , high grade glioma , Huntington's disease , long QT syndrome , lung adenocarcinoma , lung non-small cell carcinoma , lung small cell carcinoma , lung squamous cell carcinoma , melanoma , Neoplasm Metastasis , osteosarcoma , pancreatic adenocarcinoma , pancreatic cancer , pancreatic ductal adenocarcinoma , prostate cancer , Sjogren's syndrome , type 1 diabetes mellitus , urinary bladder cancer , viral encephalitis E2f3 genetic disease , high grade glioma , lung adenocarcinoma , lung non-small cell carcinoma , lung squamous cell carcinoma , prostate cancer Egf acute kidney failure , adenocarcinoma , AIDS-Associated Nephropathy , Alzheimer's disease , autistic disorder , Breast Neoplasms , cholangiocarcinoma , cleft palate , dermatitis , Diabetic Nephropathies , Disease Progression , endometrial cancer , Endometrial Neoplasms , Experimental Liver Cirrhosis , gallbladder cancer , gastric ulcer , genetic disease , Genetic Predisposition to Disease , glioblastoma , Helicobacter Infections , hepatocellular carcinoma , hereditary renal cell carcinoma , high grade glioma , inclusion body myopathy with Paget disease of bone and frontotemporal dementia , juvenile rheumatoid arthritis , kidney disease , lung adenocarcinoma , lung non-small cell carcinoma , lupus nephritis , Metaplasia , Neoplasm Invasiveness , Neoplasm Metastasis , Neoplastic Cell Transformation , oligodendroglioma , ovarian cancer , pancreatic cancer , pancreatic ductal carcinoma , pancreatitis , Parkinson's disease , perinatal necrotizing enterocolitis , pneumonia , pre-malignant neoplasm , primary hypomagnesemia , Prostatic Neoplasms , pulmonary fibrosis , renal hypomagnesemia 3 , renal hypomagnesemia 4 , renal hypomagnesemia 5 with ocular involvement , Reperfusion Injury , schizophrenia , shigellosis , Tympanic Membrane Perforation , Wounds and Injuries , Zollinger-Ellison syndrome Egfr acute kidney failure , Acute Lung Injury , adenocarcinoma , adrenocortical carcinoma , Alcoholic Liver Diseases , Alzheimer's disease , Animal Mammary Neoplasms , aortic valve disease , aortic valve stenosis , arteriovenous malformations of the brain , asthma , autistic disorder , autosomal recessive polycystic kidney disease , Bannayan-Riley-Ruvalcaba syndrome , Bile Duct Neoplasms , Brain Neoplasms , breast cancer , breast carcinoma , Breast Neoplasms , bronchiolo-alveolar adenocarcinoma , carcinoma , Cardiomegaly , cerebral palsy , Chloracne , cholangiocarcinoma , chronic obstructive pulmonary disease , Colonic Neoplasms , colorectal cancer , Colorectal Neoplasms , Coronavirus infectious disease , Craniofacial Abnormalities , dementia , dermatitis , diabetes mellitus , diabetic retinopathy , dilated cardiomyopathy , Disease Progression , endometrial carcinoma , Endometrioid Carcinomas , endometriosis , esophageal carcinoma , Esophageal Neoplasms , esophagus squamous cell carcinoma , Experimental Arthritis , Experimental Diabetes Mellitus , Experimental Mammary Neoplasms , familial adenomatous polyposis , Gallbladder Neoplasms , genetic disease , glioblastoma , head and neck cancer , Head and Neck Neoplasms , head and neck squamous cell carcinoma , Helicobacter Infections , hepatocellular carcinoma , hereditary breast ovarian cancer syndrome , Hereditary Neoplastic Syndromes , high grade glioma , Huntington's disease , Hyperplasia , hypertension , Immediate Hypersensitivity , Inflammation , Insulin Resistance , Laryngeal Neoplasms , laryngeal squamous cell carcinoma , Left Ventricular Hypertrophy , leiomyoma , liver cirrhosis , lung adenocarcinoma , lung cancer , lung carcinoma , lung disease , Lung Neoplasms , lung non-small cell carcinoma , lung small cell carcinoma , lung squamous cell carcinoma , Lymphatic Metastasis , malignant mesothelioma , median neuropathy , Mesothelioma , Nasopharyngeal Neoplasms , Neonatal Inflammatory Skin and Bowel Disease 2 , Neoplasm Invasiveness , Neoplasm Metastasis , Neoplasm Recurrence, Local , obesity , oral squamous cell carcinoma , osteosarcoma , ovarian cancer , Ovarian Neoplasms , pancreatic cancer , papilloma , papillomavirus infectious disease , Parkinson's disease , Parkinsonism , peptic esophagitis , Phyllodes Tumor , pleomorphic xanthoastrocytoma , prostate cancer , Prostatic Neoplasms , pulmonary fibrosis , pulmonary hypertension , Pulmonary Hypertension, Hypoxia-Induced , pulmonary tuberculosis , rectal benign neoplasm , renal cell carcinoma , Right Ventricular Hypertrophy , schizophrenia , severe acute respiratory syndrome , Spinal Cord Injuries , squamous cell carcinoma , stomach cancer , Stomach Neoplasms , Tobacco Use Disorder , tongue squamous cell carcinoma , transitional cell carcinoma , type 1 diabetes mellitus , type 2 diabetes mellitus , urinary bladder cancer , Uterine Cervical Neoplasms , Vascular Remodeling , Ventricular Fibrillation Erbb2 adenocarcinoma , amblyopia , Animal Disease Models , Animal Mammary Neoplasms , basal cell carcinoma , Breast Cancer, Familial , Breast Neoplasms , calcinosis , Carcinogenesis , cholangiocarcinoma , Circulating Neoplastic Cells , Colonic Neoplasms , colorectal cancer , Colorectal Neoplasms , congestive heart failure , Diabetic Cardiomyopathies , dilated cardiomyopathy , disease of cellular proliferation , Disease Progression , ductal carcinoma in situ , Endometrial Neoplasms , Endometrioid Carcinomas , esophageal carcinoma , Esophageal Neoplasms , Experimental Mammary Neoplasms , Experimental Neoplasms , Familial Visceral Neuropathy 2, Autosomal Recessive , gallbladder carcinoma , Gallbladder Neoplasms , gastric adenocarcinoma , Gastrointestinal Neoplasms , genetic disease , Germ Cell and Embryonal Neoplasms , head and neck squamous cell carcinoma , Helicobacter Infections , HELLP syndrome , high grade glioma , Hirschsprung's disease , hypertrophic cardiomyopathy 25 , interstitial cystitis , invasive ductal carcinoma , invasive lobular carcinoma , Kidney Neoplasms , laryngeal squamous cell carcinoma , lung adenocarcinoma , lung cancer , Lung Neoplasms , lung non-small cell carcinoma , Lymphatic Metastasis , mammary Paget's disease , median neuropathy , medulloblastoma , melanoma , Mouth Neoplasms , myelodysplastic syndrome , myocardial infarction , Myocardial Ischemia , Neoplasm Metastasis , Neoplasm Recurrence, Local , nephroblastoma , oral squamous cell carcinoma , ovarian cancer , ovarian carcinoma , Ovarian Neoplasms , ovarian serous cystadenocarcinoma , ovary adenocarcinoma , pancreatic adenocarcinoma , papillary renal cell carcinoma , papilloma , Parkinson's disease , pre-eclampsia , prostate adenocarcinoma , prostate cancer , Prostatic Neoplasms , Recurrence , renal cell carcinoma , skin melanoma , Sporadic Papillary Renal Cell Carcinoma , stomach cancer , stomach carcinoma , Stomach Neoplasms , transitional cell carcinoma , urinary bladder cancer , Urologic Neoplasms , uterine cancer , uterine carcinosarcoma , Uterine Cervical Neoplasms , Ventilator-Induced Lung Injury , Wallerian Degeneration Fhit amenorrhea , amphetamine abuse , autistic disorder , Breast Neoplasms , carcinoma , Endometrial Neoplasms , genetic disease , Germ Cell and Embryonal Neoplasms , granulosa cell tumor , head and neck squamous cell carcinoma , hepatocellular carcinoma , invasive ductal carcinoma , Liver Neoplasms , Lung Neoplasms , lung non-small cell carcinoma , megacolon , Mesothelioma , Muir-Torre syndrome , Ovarian Neoplasms , prostate cancer , Prostatic Neoplasms , Pulmonary Arterial Hypertension , renal cell carcinoma , schizophrenia , seminoma , squamous cell carcinoma , Stomach Neoplasms , substance-related disorder , urinary bladder cancer , Urologic Neoplasms , Uterine Cervical Neoplasms , Vulvar Neoplasms Foxo3 adenoid cystic carcinoma , Alzheimer's disease , Brain Hypoxia-Ischemia , Carotid Artery Injuries , Charcot-Marie-Tooth disease type 4 , choroid plexus carcinoma , chronic obstructive pulmonary disease , dermoid cyst of ovary , Embryo Loss , endometrial adenocarcinoma , Experimental Autoimmune Encephalomyelitis , Experimental Diabetes Mellitus , genetic disease , Habitual Abortions , hemangioma , Intimal Hyperplasia , Left Ventricular Hypertrophy , liver cirrhosis , lung adenocarcinoma , lung non-small cell carcinoma , lymphoblastic lymphoma , medulloblastoma , obesity , prostate cancer , rheumatoid arthritis , Spinal Cord Injuries , status epilepticus , temporal lobe epilepsy Grb2 breast cancer , endometrial carcinoma , genetic disease , prostate adenocarcinoma , prostate cancer Hras acute myeloid leukemia , adenoid cystic carcinoma , Alzheimer's disease , angiosarcoma , Animal Mammary Neoplasms , autistic disorder , autoimmune hepatitis , B-Cell Chronic Lymphocytic Leukemia , bladder carcinoma , Brain Neoplasms , Breast Neoplasms , carcinoma , cardiofaciocutaneous syndrome , Chanarin-Dorfman syndrome , chronic lymphocytic leukemia , Colorectal Neoplasms , complex cortical dysplasia with other brain malformations , congenital fibrosis of the extraocular muscles , Congenital Myopathy with Excess of Muscle Spindles , Costello syndrome , delta beta-thalassemia , developmental and epileptic encephalopathy , diabetic retinopathy , disease of cellular proliferation , Disease Progression , early infantile epileptic encephalopathy , endometrial carcinoma , epidermal nevus , esophageal carcinoma , Experimental Diabetes Mellitus , Experimental Liver Neoplasms , Experimental Mammary Neoplasms , Experimental Neoplasms , Follicular Thyroid Cancer , gastric adenocarcinoma , genetic disease , glioblastoma , head and neck squamous cell carcinoma , hepatocellular adenoma , hepatocellular carcinoma , Hereditary Neoplastic Syndromes , high grade glioma , Hydrops Fetalis , Hypophosphatemic Rickets , immunodeficiency 39 , intellectual disability , large congenital melanocytic nevus , Leber congenital amaurosis , linear nevus sebaceous syndrome , liver cancer , liver carcinoma , liver cirrhosis , Liver Neoplasms , lung adenocarcinoma , Lung Neoplasms , lung non-small cell carcinoma , lung squamous cell carcinoma , melanoma , Mouth Neoplasms , multiple myeloma , myelodysplastic syndrome , Nasopharyngeal Neoplasms , Neoplasm Invasiveness , Neoplastic Cell Transformation , Neoplastic Processes , neuroblastoma , neuronal ceroid lipofuscinosis , Noonan syndrome , Noonan syndrome 1 , Noonan syndrome 3 , obsessive-compulsive disorder , ovarian cancer , Ovarian Neoplasms , ovarian serous cystadenocarcinoma , pancreatic adenocarcinoma , papillary renal cell carcinoma , papilloma , Paraproteinemias , parathyroid adenoma , Penile Neoplasms , phobic disorder , Pigmented Nevus , plasma cell neoplasm , pre-malignant neoplasm , prostate adenocarcinoma , Prostate Cancer, Hereditary, 1 , Prostatic Neoplasms , pulmonary valve stenosis , RASopathy , Recurrence , rhabdomyosarcoma , Salivary Gland Neoplasms , Sebaceous Nevus Syndrome and Hemimegalencephaly , seminoma , skin benign neoplasm , skin melanoma , Skin Neoplasms , spermatocytoma , Splenic Neoplasms , Sporadic Papillary Renal Cell Carcinoma , squamous cell carcinoma , Stomach Neoplasms , T-cell non-Hodgkin lymphoma , thymoma , Thyroid Neoplasms , Tongue Neoplasms , transitional cell carcinoma , Trichilemmoma , urinary bladder cancer , Urinary Bladder Neoplasm , uterine cancer , uterine carcinosarcoma , Uterine Cervical Neoplasms Kras Acute Experimental Pancreatitis , acute lymphoblastic leukemia , acute myeloid leukemia , adenocarcinoma , adenoid cystic carcinoma , adenoma , anaplastic astrocytoma , anemia , angiosarcoma , Animal Disease Models , arteriovenous malformations of the brain , autoimmune lymphoproliferative syndrome type 4 , B-Cell Chronic Lymphocytic Leukemia , breast adenocarcinoma , breast cancer , Breast Cancer, Familial , Breast Neoplasms , bronchiolo-alveolar adenocarcinoma , Capillary Malformation-Arteriovenous Malformation 1 , Carcinogenesis , carcinoma , cardiofaciocutaneous syndrome , cardiofaciocutaneous syndrome 1 , cardiofaciocutaneous syndrome 2 , cardiomyopathy , Cecal Neoplasms , cholangiocarcinoma , chronic lymphocytic leukemia , chronic myelogenous leukemia, BCR-ABL1 positive , chronic myeloid leukemia , Colonic Neoplasms , colorectal cancer , Colorectal Neoplasms , Costello syndrome , dilated cardiomyopathy , disease of cellular proliferation , Disease Progression , Encephalocraniocutaneous Lipomatosis , endometrial adenocarcinoma , endometrial carcinoma , endometrial hyperplasia , Endometrial Hyperplasia without Atypia , Endometrial Intraepithelial Neoplasia , Endometrial Neoplasms , Endometrioid Carcinomas , endometriosis , epidermal nevus , esophageal carcinoma , Experimental Liver Neoplasms , Experimental Neoplasms , Experimental Sarcoma , fibrillary astrocytoma , gallbladder cancer , Gallbladder Neoplasms , gastric adenocarcinoma , gastrointestinal stromal tumor , genetic disease , glioblastoma , hepatocellular adenoma , hepatocellular carcinoma , hereditary breast ovarian cancer syndrome , hereditary diffuse gastric cancer , high grade glioma , Hodgkin's lymphoma , Hydrops Fetalis , hypertension , intellectual disability , intrahepatic cholangiocarcinoma , juvenile myelomonocytic leukemia , juvenile rheumatoid arthritis , Kidney Neoplasms , Klatskin's tumor , Leukocytosis , linear nevus sebaceous syndrome , Liver Neoplasms , lung adenocarcinoma , lung cancer , lung carcinoma , Lung Neoplasms , lung non-small cell carcinoma , Lung Reperfusion Injury , lung sarcomatoid carcinoma , lung squamous cell carcinoma , lymphoma , Lynch syndrome , malignant astrocytoma , melanoma , Mouth Neoplasms , Multiple Abnormalities , multiple myeloma , muscular atrophy , myelodysplastic syndrome , myeloid neoplasm , myeloid sarcoma , Neoplasm Metastasis , Neoplastic Cell Transformation , neuroblastoma , Noonan syndrome , Noonan syndrome 1 , Noonan syndrome 3 , oculoectodermal syndrome , Ovarian Neoplasms , ovary serous adenocarcinoma , pancreatic adenocarcinoma , pancreatic cancer , pancreatic carcinoma , pancreatic ductal carcinoma , pancreatic mucinous cystadenoma , Paraproteinemias , Parasitic Liver Diseases , Penile Neoplasms , pilocytic astrocytoma , plasma cell neoplasm , prostate cancer , Prostate Cancer, Hereditary, 1 , prostate carcinoma in situ , Prostatic Neoplasms , RASopathy , Recurrence , renal cell carcinoma , renal fibrosis , Sebaceous Nevus Syndrome and Hemimegalencephaly , seminoma , skin melanoma , Splenomegaly , squamous cell carcinoma , stomach cancer , stomach carcinoma , Stomach Neoplasms , T-cell acute lymphoblastic leukemia , teratoma , Thyroid Carcinoma, Nonmedullary 1 , thyroid gland follicular carcinoma , thyroid gland medullary carcinoma , Thyroid Neoplasms , transitional cell carcinoma , Triple Negative Breast Neoplasms , urinary bladder cancer , Urinary Bladder Neoplasm , uterine cancer , Uterine Cervical Neoplasms , Uterine Neoplasms , Vascular Malformations Map2k1 autism spectrum disorder , Bloom syndrome , brain ischemia , breast cancer , cardiofaciocutaneous syndrome , cardiofaciocutaneous syndrome 3 , Cardiovascular Abnormalities , cataract , colorectal cancer , Colorectal Neoplasms , congestive heart failure , Costello syndrome , disease of cellular proliferation , Experimental Liver Neoplasms , Familial Prostate Cancer , gastric adenocarcinoma , genetic disease , hairy cell leukemia , head and neck squamous cell carcinoma , high grade glioma , lung adenocarcinoma , lung non-small cell carcinoma , lung squamous cell carcinoma , melanoma , melorheostosis , neuronal ceroid lipofuscinosis , non-Hodgkin lymphoma , Noonan syndrome , Noonan syndrome 1 , ovarian carcinoma , Ovarian Neoplasms , pancreatic adenocarcinoma , pancreatitis , prostate adenocarcinoma , Prostatic Neoplasms , RASopathy , renal cell carcinoma , skin melanoma , small intestine adenocarcinoma , Sturge-Weber syndrome , transitional cell carcinoma , uterine cancer , Vascular Malformations Map2k2 adenoid cystic carcinoma , amenorrhea , breast cancer , cardiofaciocutaneous syndrome , cardiofaciocutaneous syndrome 1 , cardiofaciocutaneous syndrome 4 , colorectal cancer , Costello syndrome , gastric adenocarcinoma , genetic disease , glioblastoma , head and neck squamous cell carcinoma , hepatocellular carcinoma , hypertrophic cardiomyopathy , lung non-small cell carcinoma , melanoma , Metabolic Syndrome , migraine , multicentric Castleman disease , neurofibromatosis-Noonan syndrome , Noonan syndrome , Noonan syndrome 1 , Noonan syndrome with multiple lentigines , Ovarian Neoplasms , pancreatic adenocarcinoma , Prostatic Neoplasms , RASopathy , skin melanoma , small intestine adenocarcinoma , Smith-Magenis syndrome Mapk1 Aberrant Crypt Foci , alcohol dependence , alcohol use disorder , Alzheimer's disease , Animal Disease Models , autism spectrum disorder , brain ischemia , breast cancer , Cardiomegaly , cardiomyopathy , childhood pilocytic astrocytoma , chromosome 22q11.2 deletion syndrome, distal , cocaine abuse , cocaine dependence , Cocaine-Related Disorders , colon adenocarcinoma , colon cancer , colorectal adenocarcinoma , Colorectal Neoplasms , congenital heart disease , coronary restenosis , Coronavirus infectious disease , demyelinating disease , depressive disorder , Diabetic Cardiomyopathies , Diabetic Nephropathies , DiGeorge syndrome , Dwarfism , endometrial adenocarcinoma , endometrial carcinoma , Endometrioid Carcinomas , Experimental Colitis , Experimental Diabetes Mellitus , Experimental Liver Cirrhosis , Experimental Liver Neoplasms , Experimental Mammary Neoplasms , Experimental Radiation Injuries , extrahepatic cholestasis , Female Infertility , gastric ulcer , genetic disease , Gliosis , glomerulonephritis , Head and Neck Neoplasms , head and neck squamous cell carcinoma , Hemorrhagic Shock , hepatocellular carcinoma , Hodgkin's lymphoma , Hyperalgesia , Hypertrophy , intellectual disability , Intimal Hyperplasia , juvenile rheumatoid arthritis , kidney disease , Kidney Neoplasms , Left Ventricular Hypertrophy , Lung Neoplasms , melanoma , Memory Disorders , middle cerebral artery infarction , morphine dependence , Multiple Organ Failure , myocardial infarction , Myocardial Reperfusion Injury , Neoplasm Invasiveness , Neoplasm Metastasis , Nerve Degeneration , neuronal ceroid lipofuscinosis 6A , Noonan syndrome 13 , obesity , Ovarian Neoplasms , pancreatic adenocarcinoma , Parkinson's disease , Pneumococcal Pneumonia , portal hypertension , Postoperative Cognitive Dysfunction , prostate adenocarcinoma , pulmonary hypertension , renal cell carcinoma , renal fibrosis , Sepsis , severe acute respiratory syndrome , Sezary's disease , Shock , squamous cell carcinoma , Stomach Neoplasms , Testis Reperfusion Injury , thyroid cancer , Thyroid Neoplasms , transitional cell carcinoma , trigeminal neuralgia , type 2 diabetes mellitus , ureteral obstruction , urethral obstruction , Urinary Incontinence , Uterine Cervical Neoplasms , vascular dementia , velocardiofacial syndrome , Weissenbacher-Zweymuller syndrome , withdrawal disorder Mapk3 Aberrant Crypt Foci , alcohol dependence , alcohol use disorder , Alzheimer's disease , Animal Disease Models , Atrophy , autism spectrum disorder , autistic disorder , brain ischemia , cardiomyopathy , chromosome 16p11.2 deletion syndrome, 593-kb , chromosome 16p11.2 duplication syndrome , cocaine abuse , cocaine dependence , Cocaine-Related Disorders , colon adenocarcinoma , colorectal adenocarcinoma , Colorectal Neoplasms , Coronavirus infectious disease , coronin-1A deficiency , Developmental Disabilities , Diabetic Cardiomyopathies , endometrial adenocarcinoma , endometrial carcinoma , Endometrial Neoplasms , Endometrioid Carcinomas , episodic kinesigenic dyskinesia 1 , Experimental Colitis , Experimental Diabetes Mellitus , Experimental Liver Cirrhosis , Experimental Liver Neoplasms , Experimental Mammary Neoplasms , Experimental Radiation Injuries , extrahepatic cholestasis , Female Infertility , gastric ulcer , genetic disease , Gliosis , Glycogen Storage Disease XII , Head and Neck Neoplasms , high grade glioma , Hyperalgesia , Hypertrophy , Insulin Resistance , Kidney Neoplasms , Lung Neoplasms , Memory Disorders , middle cerebral artery infarction , morphine dependence , Myocardial Reperfusion Injury , Neoplasm Invasiveness , Neoplasm Metastasis , Nerve Degeneration , Neurodevelopmental Disorders , neuronal ceroid lipofuscinosis 6A , obesity , Osteoarthritis, Experimental , Ovarian Neoplasms , pancreatic ductal carcinoma , Parkinson's disease , Pneumococcal Pneumonia , Postoperative Cognitive Dysfunction , Prostatic Neoplasms , pulmonary hypertension , schizophrenia , Sepsis , severe acute respiratory syndrome , Shock , spondylocostal dysostosis 5 , squamous cell carcinoma , Stomach Neoplasms , Testis Reperfusion Injury , trigeminal neuralgia , type 2 diabetes mellitus , ureteral obstruction , withdrawal disorder Met acute kidney failure , Acute Liver Failure , acute myeloid leukemia , adenocarcinoma , Animal Disease Models , Animal Hepatitis , arteriosclerosis , autism spectrum disorder , autistic disorder , autosomal recessive nonsyndromic deafness 97 , Bile Duct Neoplasms , breast cancer , breast carcinoma , Carcinogenesis , carcinoma , Cardiomegaly , childhood hepatocellular carcinoma , cholangiocarcinoma , Chronic Hepatitis , cognitive disorder , colorectal cancer , Colorectal Neoplasms , congenital diaphragmatic hernia , Copper-Overload Cirrhosis , Cystic, Mucinous, and Serous Neoplasms , disease of cellular proliferation , distal arthrogryposis type 1 , Distal Arthrogryposis Type 11 , distal arthrogryposis type 1A , Endometrial Neoplasms , Esophageal Neoplasms , Experimental Diabetes Mellitus , Experimental Liver Cirrhosis , Experimental Liver Neoplasms , Gallbladder Neoplasms , gastrinoma , genetic disease , gestational diabetes , Gilles de la Tourette syndrome , glioblastoma , hepatoblastoma , hepatocellular carcinoma , Hepatomegaly , Hereditary Neoplastic Syndromes , high grade glioma , Hodgkin's lymphoma , Hypertriglyceridemia , intellectual disability , invasive ductal carcinoma , Kidney Neoplasms , large cell carcinoma , liver benign neoplasm , lung carcinoma , Lung Neoplasms , lung non-small cell carcinoma , lung squamous cell carcinoma , Lymphatic Metastasis , lymphedema , malignant astrocytoma , malignant mesothelioma , melanoma , metabolic dysfunction-associated steatotic liver disease , multiple myeloma , Nasal Polyps , Neoplasm Invasiveness , Neoplasm Metastasis , Nerve Sheath Neoplasms , neuroblastoma , nonsyndromic deafness , oral squamous cell carcinoma , Osteofibrous Dysplasia , osteosarcoma , ovarian cancer , Ovarian Neoplasms , pancreatic cancer , pancreatic ductal carcinoma , pancreatitis , papillary renal cell carcinoma , periodontitis , pleomorphic xanthoastrocytoma , primary biliary cholangitis , prostate carcinoma , Prostatic Neoplasms , renal carcinoma , renal cell carcinoma , Renal Cell Carcinoma 1 , Reperfusion Injury , sarcoma , schizophrenia , sinusitis , Spinal Cord Injuries , squamous cell carcinoma , steatotic liver disease , Stomach Neoplasms , tongue squamous cell carcinoma Nras acute lymphoblastic leukemia , acute myeloid leukemia , acute promyelocytic leukemia , adrenocortical carcinoma , adult T-cell leukemia/lymphoma , anemia , autoimmune lymphoproliferative syndrome , autoimmune lymphoproliferative syndrome type 4 , B-Cell Chronic Lymphocytic Leukemia , B-cell lymphoma , Brain Neoplasms , cervical cancer , cholangitis , chronic lymphocytic leukemia , chronic myelogenous leukemia, BCR-ABL1 positive , chronic myeloid leukemia , chronic myelomonocytic leukemia , colon carcinoma , colorectal cancer , Colorectal Neoplasms , disease of cellular proliferation , epidermal nevus , Experimental Arthritis , Experimental Leukemia , Experimental Liver Neoplasms , Experimental Neoplasms , Experimental Sarcoma , focal epilepsy , Follicular Thyroid Cancer , gastric adenocarcinoma , genetic disease , glioblastoma , hepatocellular carcinoma , hereditary spastic paraplegia 47 , juvenile myelomonocytic leukemia , large congenital melanocytic nevus , leukemia , Leukocytosis , linear nevus sebaceous syndrome , liver carcinoma , Liver Metastasis , lung adenocarcinoma , lung non-small cell carcinoma , lymphoma , medulloblastoma , melanoma , Mouth Neoplasms , multiple myeloma , myelodysplastic syndrome , myeloid leukemia associated with Down Syndrome , myeloid neoplasm , Nasopharyngeal Neoplasms , neuroblastoma , Neurocutaneous Melanosis , Neurodevelopmental Disorders , neurofibrosarcoma , non-Hodgkin lymphoma , Noonan syndrome , Noonan syndrome 1 , Noonan syndrome 6 , ovarian serous cystadenocarcinoma , Paraproteinemias , Pigmented Nevus , plasma cell neoplasm , prostate cancer , Prostate Cancer, Hereditary, 1 , Pyogenic Granuloma , RASopathy , rectal benign neoplasm , renal cell carcinoma , Renal Cell Carcinoma 1 , Sebaceous Nevus Syndrome and Hemimegalencephaly , skin melanoma , Splenomegaly , Stomach Neoplasms , Thyroid Neoplasms , transitional cell carcinoma , uterine cancer Pdpk1 Breast Neoplasms , congestive heart failure , diffuse large B-cell lymphoma , epilepsy , genetic disease , idiopathic generalized epilepsy , lung non-small cell carcinoma , Lymphatic Metastasis , prostate adenocarcinoma , prostate cancer , short-rib thoracic dysplasia 9 with or without polydactyly Pik3ca adenocarcinoma , adenoid cystic carcinoma , adrenocortical carcinoma , Agenesis of Corpus Callosum , autosomal recessive polycystic kidney disease , Bannayan-Riley-Ruvalcaba syndrome , basal cell carcinoma , Brain Neoplasms , breast adenocarcinoma , breast angiosarcoma , breast cancer , Breast Cancer, Familial , breast carcinoma , Breast Neoplasms , Cardiovascular Abnormalities , Cerebral Cavernous Malformation 4 , CLAPO Syndrome , CLOVES syndrome , colon adenocarcinoma , colon carcinoma , colorectal cancer , colorectal carcinoma , Colorectal Neoplasms , complex cortical dysplasia with other brain malformations , Cowden syndrome , Cowden syndrome 5 , Currarino syndrome , cutaneous Paget's disease , Developmental Disabilities , Diabetic Cardiomyopathies , diaphragmatic eventration , diffuse large B-cell lymphoma , disease of cellular proliferation , Disease Progression , Down syndrome , endometrial carcinoma , Endometrial Neoplasms , epidermal nevus , esophageal cancer , esophageal carcinoma , esophagus adenocarcinoma , estrogen-receptor positive breast cancer , Facial Asymmetry , familial multiple nevi flammei , gallbladder cancer , gallbladder carcinoma , gastric adenocarcinoma , Gastrointestinal Neoplasms , genetic disease , glioblastoma , glycogen storage disease II , Growth Disorders , head and neck cancer , Head and Neck Neoplasms , head and neck squamous cell carcinoma , Hemimegalencephaly , hepatocellular carcinoma , Hereditary Neoplastic Syndromes , high grade glioma , Hyperplasia , Hypertelorism , keratoacanthoma , Klippel-Trenaunay syndrome , Leber congenital amaurosis , liver cancer , lung adenocarcinoma , lung cancer , lung carcinoma , Lung Neoplasms , lung non-small cell carcinoma , lung oat cell carcinoma , lung small cell carcinoma , lung squamous cell carcinoma , Lymphatic Metastasis , Macrocephaly , medulloblastoma , Megalencephaly - Cutis Marmorata Telangiectatica Congenita , Megalencephaly-Polymicrogyria-Polydactyly-Hydrocephalus Syndrome , Megalodactyly , melanoma , Mouth Neoplasms , multiple myeloma , Nasopharyngeal Neoplasms , Neoplasm Metastasis , neuroblastoma , NEURODEVELOPMENTAL DISORDER WITH MICROCEPHALY, ARTHROGRYPOSIS, AND STRUCTURAL BRAIN ANOMALIES , non-Hodgkin lymphoma , Noonan syndrome 8 , ovarian cancer , Ovarian Neoplasms , ovarian serous cystadenocarcinoma , pancreatic adenocarcinoma , papillary renal cell carcinoma , Paraproteinemias , Penile Neoplasms , plasma cell neoplasm , polycystic kidney disease , prostate adenocarcinoma , prostate cancer , Prostatic Neoplasms , PTEN hamartoma tumor syndrome , rectal benign neoplasm , renal cell carcinoma , Renal Cell Carcinoma 1 , rosette-forming glioneuronal tumor , sarcoma , seborrheic keratosis , skin melanoma , Sporadic Papillary Renal Cell Carcinoma , squamous cell carcinoma , stomach cancer , Stomach Neoplasms , Stroke , Thyroid Neoplasms , transitional cell carcinoma , urinary bladder cancer , uterine cancer , uterine carcinosarcoma , Uterine Cervical Neoplasms , Vascular Malformations , Ventricular Dysfunction, Left , Weight Gain Pik3cb colorectal cancer , diffuse large B-cell lymphoma , endometrial cancer , Endometrial Neoplasms , genetic disease , glioblastoma , hepatocellular carcinoma , Lymphatic Metastasis , NATURAL KILLER CELL ENTEROPATHY , prostate adenocarcinoma , prostate cancer , Prostatic Neoplasms , rectal benign neoplasm , schizophrenia , Subarachnoid Hemorrhage , thrombosis , type 2 diabetes mellitus Pik3cd Charcot-Marie-Tooth disease type 2 , Chlamydia pneumonia , chromosome 1p36 deletion syndrome , colorectal cancer , cutaneous leishmaniasis , diffuse large B-cell lymphoma , endometrial cancer , Experimental Arthritis , genetic disease , glioblastoma , immunodeficiency 14 , listeriosis , mantle cell lymphoma , Neurodevelopmental Disorders , primary immunodeficiency disease , Prostatic Neoplasms , Roifman-Chitayat Syndrome , schizophrenia , Streptococcus pneumonia , toxic shock syndrome , visceral leishmaniasis Pik3r1 agammaglobulinemia 7 , Alzheimer's disease , Animal Mammary Neoplasms , astroblastoma , Brain Neoplasms , Breast Neoplasms , Burkitt lymphoma , carcinoma , CLOVES syndrome , Colonic Neoplasms , colorectal cancer , Colorectal Neoplasms , disease of cellular proliferation , endometrial adenocarcinoma , endometrial cancer , endometrial carcinoma , Experimental Diabetes Mellitus , Experimental Mammary Neoplasms , genetic disease , glioblastoma , glucose intolerance , hepatocellular carcinoma , hypertension , hypoglycemia , immunodeficiency 36 , Insulin Resistance , lung adenocarcinoma , lung small cell carcinoma , Megalencephaly - Cutis Marmorata Telangiectatica Congenita , melanoma , Nematode Infections , Neurodevelopmental Disorders , obesity , osteoarthritis , Ovarian Neoplasms , portal hypertension , primary immunodeficiency disease , prostate cancer , Prostatic Neoplasms , renal cell carcinoma , Reperfusion Injury , rheumatoid arthritis , Septic Peritonitis , SHORT syndrome , skin melanoma , Smith-Kingsmore Syndrome , type 2 diabetes mellitus , uterine cancer , uterine carcinosarcoma , Vascular Malformations , vitiligo , X-linked agammaglobulinemia Pik3r2 colon cancer , Colonic Neoplasms , Developmental Disabilities , Endometrioid Carcinomas , epilepsy , genetic disease , hepatocellular carcinoma , intellectual disability , Megalencephaly - Cutis Marmorata Telangiectatica Congenita , Megalencephaly-Polymicrogyria-Polydactyly-Hydrocephalus Syndrome , Megalencephaly-Polymicrogyria-Polydactyly-Hydrocephalus Syndrome 1 , multiple myeloma , pancreatic ductal adenocarcinoma , prostate cancer , Prostatic Neoplasms , Smith-Kingsmore Syndrome Pik3r3 Breast Cancer, Familial , Charcot-Marie-Tooth disease dominant intermediate C , colon adenocarcinoma , Colorectal Neoplasms , developmental and epileptic encephalopathy , early infantile epileptic encephalopathy , genetic disease , hepatocellular carcinoma , high grade glioma , Neoplasm Metastasis Pik3r5 ataxia with oculomotor apraxia type 3 , colon cancer , genetic disease , spastic ataxia Plcg1 acute myocardial infarction , adult T-cell leukemia/lymphoma , Alzheimer's disease , angiosarcoma , breast cancer , breast carcinoma , colon cancer , colorectal cancer , Experimental Diabetes Mellitus , familial adenomatous polyposis , focal epilepsy , genetic disease , hepatocellular carcinoma , hereditary breast ovarian cancer syndrome , IMMUNE DYSREGULATION, AUTOIMMUNITY, AND AUTOINFLAMMATION , laryngeal squamous cell carcinoma , lung adenocarcinoma , lung cancer , lung non-small cell carcinoma , oral squamous cell carcinoma , polycystic kidney disease , primary cutaneous T-cell non-Hodgkin lymphoma , Sezary's disease Plcg2 Alzheimer's disease , Autoinflammation, Antibody Deficiency, and Immune Dysregulation, PLCG2-Associated , B-Cell Chronic Lymphocytic Leukemia , Cryopyrin-Associated Periodic Syndromes , familial cold autoinflammatory syndrome , familial cold autoinflammatory syndrome 3 , Fever , genetic disease , inflammatory bowel disease Prkca brain ischemia , Breast Neoplasms , Cardiomegaly , chordoid glioma , congestive heart failure , dilated cardiomyopathy , ductal carcinoma in situ , endometrial carcinoma , Experimental Diabetes Mellitus , genetic disease , high grade glioma , hypertension , Intestinal Neoplasms , ischemia , large cell carcinoma , Left Ventricular Hypertrophy , metabolic dysfunction-associated steatotic liver disease , nephrogenic diabetes insipidus , occupational asthma , ovarian cancer , ovarian carcinoma , Pituitary Neoplasms , Polyuria , prostate carcinoma , Reperfusion Injury , Right Ventricular Hypertrophy , urinary bladder cancer Prkcb adult T-cell leukemia/lymphoma , Alzheimer's disease , autistic disorder , Cardiomegaly , Colorectal Neoplasms , congestive heart failure , Diabetic Nephropathies , dilated cardiomyopathy , Experimental Diabetes Mellitus , Experimental Liver Cirrhosis , Fetal Diseases , genetic disease , Hyperalgesia , hyperglycemia , hypertension , immune system disease , Insulin Resistance , Left Ventricular Hypertrophy , lung non-small cell carcinoma , Myocardial Reperfusion Injury , obesity , sensorineural hearing loss , silicosis , status epilepticus , Stomach Neoplasms , type 2 diabetes mellitus Prkcg Animal Disease Models , autosomal dominant cerebellar ataxia , autosomal recessive spinocerebellar ataxia 14 , cerebellar ataxia , familial cold autoinflammatory syndrome 2 , genetic disease , hereditary ataxia , Hyperalgesia , intellectual disability , middle cerebral artery infarction , primary cerebellar degeneration , retinitis pigmentosa , Spinal Cord Injuries , spinocerebellar ataxia type 14 , Spinocerebellar Ataxias , trigeminal neuralgia Pten acute lymphoblastic leukemia , acute megakaryocytic leukemia , acute myocardial infarction , adenocarcinoma , adenoid cystic carcinoma , angiomyolipoma , anisometropia , asthma , Ataxia , autism spectrum disorder , autistic disorder , bacterial pneumonia , Bannayan-Riley-Ruvalcaba syndrome , biliary atresia , bladder squamous cell carcinoma , brain disease , Brain Neoplasms , breast cancer , Breast Cancer, Familial , breast carcinoma , Breast Neoplasms , carcinoma , Cardiomegaly , Cardiovascular Abnormalities , cataract , cerebral cavernous malformation , cervix uteri carcinoma in situ , Chlamydiaceae Infections , cholangiocarcinoma , colon cancer , colon carcinoma , colorectal cancer , Colorectal Neoplasms , Congenital Hypomyelinating Neuropathy , Cowden syndrome , Cowden syndrome 4 , Craniofacial Abnormalities , developmental coordination disorder , Developmental Disabilities , diabetes mellitus , Diabetic Nephropathies , diabetic retinopathy , diffuse large B-cell lymphoma , disease of cellular proliferation , Disease Progression , Ductal Carcinoma , endometrial adenocarcinoma , endometrial cancer , endometrial carcinoma , endometrial hyperplasia , Endometrial Intraepithelial Neoplasia , Endometrial Neoplasms , Endometrioid Carcinomas , epilepsy , Epstein-Barr virus infectious disease , esophageal cancer , Experimental Mammary Neoplasms , Experimental Melanoma , familial meningioma , fetal alcohol spectrum disorder , Follicular Thyroid Cancer , gastric adenocarcinoma , Gastrointestinal Neoplasms , Generalized Epilepsy , genetic disease , glioblastoma , head and neck squamous cell carcinoma , Helicobacter Infections , hemangioma , hemangiopericytoma , Hemimegalencephaly , hepatitis B , hepatocellular adenoma , hepatocellular carcinoma , Hepatomegaly , hereditary breast ovarian cancer syndrome , Hereditary Neoplastic Syndromes , high grade glioma , hyperinsulinism , hypertension , Hypertrophy , in situ carcinoma , Insulin Resistance , intellectual disability , intermediate coronary syndrome , intestinal pseudo-obstruction , Intimal Hyperplasia , intrahepatic cholangiocarcinoma , juvenile polyposis syndrome , Language Development Disorders , Left Ventricular Hypertrophy , leiomyoma , leukemia , lipoma , lipomatosis , liver cirrhosis , long QT syndrome , lung adenocarcinoma , Lung Neoplasms , lung non-small cell carcinoma , lung oat cell carcinoma , lung small cell carcinoma , lung squamous cell carcinoma , Lymphatic Metastasis , lymphoma , Macrocephaly , macrocephaly-autism syndrome , major depressive disorder , melanoma , meningioma , metabolic dysfunction-associated steatotic liver disease , Mouth Neoplasms , Multiple Abnormalities , muscular dystrophy , myeloproliferative neoplasm , Myocardial Reperfusion Injury , Neoplasm Invasiveness , Neoplasm Metastasis , Neoplastic Cell Transformation , Nerve Sheath Neoplasms , Nervous System Malformations , Neurodevelopmental Disorders , Noonan syndrome with multiple lentigines , oral squamous cell carcinoma , osteochondrodysplasia , ovarian cancer , Ovarian Neoplasms , ovarian serous carcinoma , pancreatic cancer , papillary tumor of the pineal region , papillomavirus infectious disease , peripheral nervous system neoplasm , persistent fetal circulation syndrome , Pneumococcal Pneumonia , polydactyly , portal hypertension , prostate adenocarcinoma , prostate cancer , Prostate Cancer, Hereditary, 1 , Prostate Cancer, Somatic , prostate carcinoma in situ , prostatic hypertrophy , Prostatic Neoplasms , Proteus syndrome , Pseudomonas Infections , PTEN hamartoma tumor syndrome , renal cell carcinoma , Renal Cell Carcinoma 1 , Reperfusion Injury , retinitis pigmentosa , rhabdomyosarcoma , schizophrenia , Septic Peritonitis , Sezary's disease , silicosis , skin disease , skin melanoma , Skin Neoplasms , skin squamous cell carcinoma , Spinal Cord Injuries , sporadic breast cancer , squamous cell carcinoma , steatotic liver disease , stomach cancer , stomach disease , substance-related disorder , thyroid gland follicular carcinoma , Thyroid Neoplasms , transient cerebral ischemia , transitional cell carcinoma , urinary bladder cancer , uterine cancer , uterine carcinosarcoma , Uterine Cervical Neoplasms , uterine corpus cancer , visual epilepsy , X-linked VACTERL association Raf1 Breast Neoplasms , Cardiomegaly , colorectal adenocarcinoma , colorectal adenoma , Colorectal Neoplasms , dilated cardiomyopathy , dilated cardiomyopathy 1B , dilated cardiomyopathy 1NN , Dilated Cardiomyopathy with Left Ventricular Noncompaction , familial hypertrophic cardiomyopathy , gastric adenocarcinoma , genetic disease , high grade glioma , Hyperalgesia , hypertrophic cardiomyopathy , hypertrophic cardiomyopathy 1 , ischemia , Kidney Neoplasms , Liver Neoplasms , lung adenocarcinoma , Lung Neoplasms , lung non-small cell carcinoma , Lymphangiectasis , malignant mesothelioma , melanoma , Noonan syndrome , Noonan syndrome 1 , Noonan syndrome 3 , Noonan syndrome 5 , Noonan syndrome with multiple lentigines , Noonan syndrome with multiple lentigines 2 , ovarian carcinoma , pancreatic adenocarcinoma , pancreatic carcinoma , Pituitary Stalk Interruption Syndrome , prostate adenocarcinoma , prostate cancer , Prostatic Neoplasms , RASopathy , skin melanoma , Stroke , Ventricular Tachycardia Rarb adenocarcinoma , asbestos-related lung carcinoma , autism spectrum disorder , autosomal dominant intellectual developmental disorder 48 , Breast Neoplasms , coloboma , endometriosis , genetic disease , Head and Neck Neoplasms , lung adenocarcinoma , Lung Neoplasms , lung non-small cell carcinoma , lung small cell carcinoma , microphthalmia , NGLY1-deficiency , sciatic neuropathy , squamous cell carcinoma , Stomach Neoplasms , syndromic microphthalmia 12 , Uterine Cervical Neoplasms Rassf1 adenocarcinoma , Animal Mammary Neoplasms , bladder disease , breast cancer , Breast Neoplasms , cervical cancer , developmental and epileptic encephalopathy , early infantile epileptic encephalopathy , endometrial carcinoma , genetic disease , high grade glioma , hypertrophic cardiomyopathy , inflammatory bowel disease , Kidney Neoplasms , Lung Neoplasms , lung non-small cell carcinoma , medulloblastoma , Mesothelioma , Micronuclei, Chromosome-Defective , neuroblastoma , ovarian cancer , Ovarian Neoplasms , primary ciliary dyskinesia , prostate cancer , Prostatic Neoplasms , renal cell carcinoma , schistosomiasis , small cell carcinoma , transitional cell carcinoma , urinary bladder cancer Rassf5 autistic disorder , familial adult myoclonic epilepsy 5 , gastrointestinal stromal tumor , genetic disease , Hypokalemic Periodic Paralysis, Type 1 , Kidney Neoplasms , lung non-small cell carcinoma , neuroblastoma , parathyroid carcinoma , systemic lupus erythematosus Rb1 Acute Erythroleukemia , acute kidney failure , acute lymphoblastic leukemia , Adrenal Gland Neoplasms , adrenocortical carcinoma , Animal Mammary Neoplasms , Autosomal Recessive Woolly Hair , B-lymphoblastic leukemia/lymphoma , B-lymphoblastic leukemia/lymphoma with ETV6-RUNX1 , basal cell carcinoma , bone osteosarcoma , brain ischemia , Breast Neoplasms , carcinoma , Cardiomegaly , cervix uteri carcinoma in situ , chromosome 13q14 deletion syndrome , disease of cellular proliferation , ductal carcinoma in situ , embryonal carcinoma , Endometrial Neoplasms , esophagus squamous cell carcinoma , Experimental Liver Neoplasms , Experimental Mammary Neoplasms , Experimental Neoplasms , familial retinoblastoma , gastrointestinal stromal tumor , genetic disease , glioblastoma , glioblastoma proneural subtype , head and neck squamous cell carcinoma , hepatocellular carcinoma , Hereditary Neoplastic Syndromes , hereditary nonpolyposis colorectal cancer type 4 , hypotrichosis 8 , in situ carcinoma , Insulin Resistance , intellectual disability , intestinal pseudo-obstruction , lung adenocarcinoma , lung non-small cell carcinoma , lung oat cell carcinoma , lung small cell carcinoma , medulloblastoma , mitochondrial DNA depletion syndrome 5 , Neoplasm Metastasis , obesity , oligodendroglioma , osteosarcoma , ovarian cancer , Ovarian Neoplasms , pancreatic cancer , Phyllodes Tumor , Pituitary Neoplasms , prostate cancer , Prostatic Neoplasms , renal cell carcinoma , retinoblastoma , seminoma , Sezary's disease , small cell carcinoma , squamous cell carcinoma , Stevens-Johnson syndrome , trilateral retinoblastoma , urinary bladder cancer , Urinary Bladder Neoplasm Rxra alcoholic cardiomyopathy , amyotrophic lateral sclerosis type 1 , congenital heart disease , developmental and epileptic encephalopathy 14 , Diaphragmatic Hernia , Ehlers-Danlos syndrome classic type 1 , Experimental Liver Cirrhosis , Experimental Liver Neoplasms , Experimental Mammary Neoplasms , genetic disease , hepatocellular carcinoma , Kleefstra syndrome 1 , Leigh disease , malignant mesothelioma , multiple myeloma , Neoplasm Metastasis , Neurogenic Inflammation , obesity , pancreatic adenocarcinoma , pancreatic cancer , pancreatic ductal carcinoma , primary coenzyme Q10 deficiency 7 , prostate carcinoma in situ , Prostatic Neoplasms , Rafiq syndrome , sciatic neuropathy , Sepsis , systemic lupus erythematosus , Thyroid Neoplasms , transitional cell carcinoma , tuberous sclerosis 1 , Wallerian Degeneration Rxrb autosomal dominant intellectual developmental disorder 5 , Breast Neoplasms , Disease Progression , genetic disease , proteasome-associated autoinflammatory syndrome 1 , Stomach Neoplasms Rxrg gastrointestinal stromal tumor , genetic disease , lung non-small cell carcinoma , Myocardial Ischemia , parathyroid carcinoma , thyroid gland papillary carcinoma Sos1 arrhythmogenic right ventricular cardiomyopathy , atrial heart septal defect , Atrial Septal Defect, Secundum Type , brain ischemia , Brugada syndrome , Cafe au lait Spots, Multiple , cardiomyopathy , cerebral infarction , cervical cancer , congenital heart disease , dilated cardiomyopathy , Dwarfism , familial hypertrophic cardiomyopathy , genetic disease , gingival fibromatosis , Gingival Fibromatosis 1 , hypertrophic cardiomyopathy , hypertrophic cardiomyopathy 1 , intellectual disability , Joubert syndrome 3 , lung adenocarcinoma , Lynch syndrome , Muscle Hypotonia , Neurodevelopmental Disorders , Noonan Like Syndrome , Noonan syndrome , Noonan syndrome 1 , Noonan syndrome 3 , Noonan syndrome 4 , Nuchal Bleb, Familial , prostate cancer , ptosis , pulmonary valve stenosis , RASopathy , Stroke , Sudden Unexpected Nocturnal Death Syndrome , uterine cancer , Ventricular Tachycardia Sos2 Brain-Lung-Thyroid Syndrome , Developmental Disease , genetic disease , L-2-hydroxyglutaric aciduria , Noonan syndrome , Noonan syndrome 1 , Noonan syndrome 9 , RASopathy Stk4 bipolar disorder , focal epilepsy , genetic disease , primary immunodeficiency disease , severe combined immunodeficiency , T-Cell Immunodeficiency, Recurrent Infections, and Autoimmunity with or without Cardiac Malformations Tgfa acute kidney failure , Animal Mammary Neoplasms , Experimental Liver Neoplasms , Experimental Mammary Neoplasms , gallbladder carcinoma , gastroenteritis , genetic disease , Genetic Predisposition to Disease , Head and Neck Neoplasms , hepatoblastoma , hepatocellular carcinoma , Inflammation , insulinoma , Knee Osteoarthritis , Liver Neoplasms , Lung Injury , lung non-small cell carcinoma , mucositis , Nerve Degeneration , nonsyndromic deafness , pancreatic cancer , papilloma , Prostatic Neoplasms , psoriasis , pulmonary edema , pulmonary fibrosis , pulmonary hypertension , Right Ventricular Hypertrophy , Stomach Neoplasms , Stroke Tp53 Acute Erythroleukemia , acute kidney failure , acute lymphoblastic leukemia , acute megakaryocytic leukemia , acute myeloid leukemia , adenocarcinoma , adenoid cystic carcinoma , adenoma , adenosine deaminase deficiency , Adrenal Cortex Neoplasms , Adrenal Gland Neoplasms , adrenocortical carcinoma , adult T-cell leukemia/lymphoma , Alzheimer's disease , amyotrophic lateral sclerosis , anaplastic astrocytoma , angiosarcoma , Animal Disease Models , Arsenic Poisoning , atherosclerosis , atypical teratoid rhabdoid tumor , Autosomal Recessive Dyskeratosis Congenita , autosomal recessive dyskeratosis congenita 3 , B-Cell Chronic Lymphocytic Leukemia , B-lymphoblastic leukemia/lymphoma , Balkan nephropathy , basal cell carcinoma , bone development disease , Bone Marrow Failure Syndrome 5 , Bone Marrow Neoplasms , bone osteosarcoma , brain ischemia , Brain Neoplasms , breast adenocarcinoma , breast cancer , Breast Cancer, Familial , breast carcinoma , Breast Neoplasms , bronchiolo-alveolar adenocarcinoma , bronchopulmonary dysplasia , Calcification of Aortic Valve , calcinosis , carcinoma , cardiomyopathy , Cardiotoxicity , carotid artery disease , cataract , Cecal Neoplasms , Central Nervous System Neoplasms , cervical adenocarcinoma , cervical cancer , cervical squamous cell carcinoma , CHARGE syndrome , Chemotherapy-Induced Febrile Neutropenia , cholangiocarcinoma , cholesteatoma of middle ear , choroid plexus carcinoma , choroid plexus papilloma , Chromosome 17 Deletion , chronic lymphocytic leukemia , chronic obstructive pulmonary disease , clear cell adenocarcinoma , clear cell renal cell carcinoma , Cognitive Dysfunction , colon carcinoma , Colon Diverticulum , Colonic Neoplasms , colorectal adenocarcinoma , colorectal cancer , colorectal carcinoma , Colorectal Neoplasms , combined oxidative phosphorylation deficiency 11 , common variable immunodeficiency , congenital fibrosarcoma , congenital merosin-deficient muscular dystrophy 1A , congenital myasthenic syndrome 2A , congestive heart failure , coronary artery disease , coronary restenosis , Craniofacial Abnormalities , demyelinating disease , Diabetic Nephropathies , Diamond-Blackfan anemia , diffuse large B-cell lymphoma , diffuse pediatric-type high-grade glioma, H3-wildtype and IDH-wildtype , DiGeorge syndrome , dilated cardiomyopathy , disease of cellular proliferation , Disease Progression , Drug-induced Neutropenia , Drug-Related Side Effects and Adverse Reactions , Ductal Carcinoma , ductal carcinoma in situ , Dwarfism , dyskeratosis congenita , Emphysema , endometrial adenocarcinoma , endometrial cancer , endometrial carcinoma , Endometrial Intraepithelial Neoplasia , Endometrial Neoplasms , esophageal cancer , esophageal carcinoma , Esophageal Neoplasms , esophagus adenocarcinoma , esophagus squamous cell carcinoma , Experimental Diabetes Mellitus , Experimental Liver Neoplasms , Experimental Mammary Neoplasms , Experimental Neoplasms , Eye Neoplasms , familial melanoma , Female Infertility , Funnel Chest , gallbladder cancer , gallbladder carcinoma , Gallbladder Neoplasms , gastric adenocarcinoma , glioblastoma , graft-versus-host disease , Graves' disease , head and neck cancer , Head and Neck Neoplasms , head and neck squamous cell carcinoma , hematologic cancer , hepatoblastoma , hepatocellular carcinoma , Hereditary Adrenocortical Carcinoma , hereditary breast ovarian cancer syndrome , Hereditary Neoplastic Syndromes , high grade glioma , hypertension , hypertrophic cardiomyopathy , Hypogonadism and Testicular Atrophy , ischemia , Kaposi's sarcoma , keratosis , Leber hereditary optic neuropathy , leukemia , Li-Fraumeni syndrome , Li-Fraumeni syndrome 1 , Li-Fraumeni-Like Syndrome , Lip Neoplasms , Liver Neoplasms , low tension glaucoma , lung adenocarcinoma , Lung Neoplasms , lung non-small cell carcinoma , lung oat cell carcinoma , lung sarcomatoid carcinoma , lung small cell carcinoma , lung squamous cell carcinoma , Lymphatic Metastasis , lymphoma , male infertility , malignant astrocytoma , malignant mesothelioma , marginal zone lymphoma , medulloblastoma , melanoma , meninges sarcoma , Mesothelioma , mismatch repair cancer syndrome , mitochondrial metabolism disease , Mouth Neoplasms , multiple myeloma , myelodysplastic syndrome , myelofibrosis , myeloid leukemia associated with Down Syndrome , myeloid neoplasm , myocardial infarction , Myocardial Reperfusion Injury , nasal cavity cancer , Nasopharyngeal Neoplasms , nasopharynx carcinoma , Neoplasm Metastasis , Neoplastic Cell Transformation , nephroblastoma , Nerve Sheath Neoplasms , nervous system disease , neuroblastoma , non-Hodgkin lymphoma , Odontogenic Myxoma , Oral Lichen Planus , oral mucosa leukoplakia , oral squamous cell carcinoma , oral submucous fibrosis , Oropharyngeal Neoplasms , osteosarcoma , ovarian cancer , ovarian carcinoma , Ovarian Neoplasms , ovarian serous cystadenocarcinoma , ovarian small cell carcinoma , ovary serous adenocarcinoma , pancreatic adenocarcinoma , pancreatic cancer , pancreatic carcinoma , pancreatic ductal carcinoma , pancreatic intraductal papillary-mucinous neoplasm , pancytopenia , papillary renal cell carcinoma , paranasal sinus benign neoplasm , Paraproteinemias , Parasitic Liver Diseases , Pediatric Adrenocortical Carcinoma , Penile Neoplasms , Phyllodes Tumor , plasma cell neoplasm , pleomorphic xanthoastrocytoma , Polyploidy , pre-malignant neoplasm , Premature Aging , primary cutaneous T-cell non-Hodgkin lymphoma , primary open angle glaucoma , prostate adenocarcinoma , prostate cancer , Prostate Cancer, Hereditary, 1 , Prostatic Neoplasms , psoriasis , pterygium , pulmonary hypertension , renal cell carcinoma , Renal Cell Carcinoma 1 , Reperfusion Injury , rhabdomyosarcoma , SAPHO syndrome , sarcoma , schizophrenia , serous cystadenocarcinoma , Sezary's disease , skin melanoma , Skin Neoplasms , small cell carcinoma , Spinal Cord Injuries , spondyloarthropathy , Sporadic Papillary Renal Cell Carcinoma , squamous cell carcinoma , stomach cancer , Stomach Neoplasms , T-cell non-Hodgkin lymphoma , thymoma , Thymus Neoplasms , thyroid gland anaplastic carcinoma , thyroid gland papillary carcinoma , Thyroid Neoplasms , Tongue Neoplasms , tongue squamous cell carcinoma , transitional cell carcinoma , transthyretin amyloidosis , traumatic brain injury , Trigeminal Nerve Injuries , ulcerative colitis , urinary bladder cancer , Urinary Bladder Neoplasm , Urogenital Neoplasms , Urologic Neoplasms , uterine cancer , uterine carcinosarcoma , Uterine Cervical Neoplasms , vascular dementia , very long chain acyl-CoA dehydrogenase deficiency , Vulvar Lichen Sclerosus , Vulvar Neoplasms , xeroderma pigmentosum
Aberrant Crypt Foci Mapk1 , Mapk3 Acute Erythroleukemia Rb1 , Tp53 Acute Experimental Pancreatitis Kras acute kidney failure Bad , Egf , Egfr , Met , Rb1 , Tgfa , Tp53 Acute Liver Failure Casp9 , Met Acute Lung Injury Akt1 , Egfr acute lymphoblastic leukemia Ccnd1 , Cdk6 , Cdkn2a , Kras , Nras , Pten , Rb1 , Tp53 acute megakaryocytic leukemia Pten , Tp53 acute myeloid leukemia Ccnd1 , Cdk6 , Cdkn2a , Hras , Kras , Met , Nras , Tp53 acute myocardial infarction Plcg1 , Pten acute promyelocytic leukemia Akt1 , Nras adenocarcinoma Akt1 , Akt2 , Ccnd1 , Cdkn2a , Egf , Egfr , Erbb2 , Kras , Met , Pik3ca , Pten , Rarb , Rassf1 , Tp53 adenoid cystic carcinoma Ccnd1 , Foxo3 , Hras , Kras , Map2k2 , Pik3ca , Pten , Tp53 adenoma Kras , Tp53 adenosine deaminase deficiency Tp53 Adrenal Cortex Neoplasms Tp53 Adrenal Gland Neoplasms Cdkn2a , Rb1 , Tp53 adrenocortical carcinoma Braf , Cdkn2a , Egfr , Nras , Pik3ca , Rb1 , Tp53 adult T-cell leukemia/lymphoma Nras , Plcg1 , Prkcb , Tp53 agammaglobulinemia 7 Pik3r1 Agenesis of Corpus Callosum Pik3ca Aicardi-Goutieres Syndrome 3 Ccnd1 AIDS-Associated Nephropathy Egf alcohol dependence Mapk1 , Mapk3 alcohol use disorder Mapk1 , Mapk3 alcoholic cardiomyopathy Rxra Alcoholic Liver Diseases Egfr Alzheimer's disease Akt1 , Bad , Casp9 , E2f1 , Egf , Egfr , Foxo3 , Hras , Mapk1 , Mapk3 , Pik3r1 , Plcg1 , Plcg2 , Prkcb , Tp53 amblyopia Erbb2 ameloblastoma Braf amenorrhea Fhit , Map2k2 amphetamine abuse Akt1 , Fhit amyotrophic lateral sclerosis Akt1 , Bad , Casp9 , Tp53 amyotrophic lateral sclerosis type 1 Rxra anaplastic astrocytoma Kras , Tp53 anemia Cdk6 , Kras , Nras angiomyolipoma Pten angiosarcoma Ccnd1 , Hras , Kras , Plcg1 , Tp53 Animal Disease Models Akt1 , Erbb2 , Kras , Mapk1 , Mapk3 , Met , Prkcg , Tp53 Animal Hepatitis Met Animal Mammary Neoplasms Ccnd1 , Cdk4 , Egfr , Erbb2 , Hras , Pik3r1 , Rassf1 , Rb1 , Tgfa anisometropia Pten Aortic Aneurysm, Giant Congenital Braf aortic valve disease Egfr aortic valve stenosis Egfr arrhythmogenic right ventricular cardiomyopathy Sos1 Arsenic Poisoning Tp53 arteriosclerosis Met arteriovenous malformations of the brain Braf , Egfr , Kras asbestos-related lung carcinoma Rarb asthma Egfr , Pten astroblastoma Pik3r1 Ataxia Pten ataxia telangiectasia Braf ataxia with oculomotor apraxia type 3 Pik3r5 atherosclerosis Akt1 , Ccnd1 , Tp53 atrial fibrillation Cdk6 atrial heart septal defect Sos1 Atrial Septal Defect, Secundum Type Sos1 Atrophy Mapk3 atypical teratoid rhabdoid tumor Tp53 autism spectrum disorder Map2k1 , Mapk1 , Mapk3 , Met , Pten , Rarb autistic disorder Egf , Egfr , Fhit , Hras , Mapk3 , Met , Prkcb , Pten , Rassf5 autoimmune hepatitis Hras autoimmune lymphoproliferative syndrome Nras autoimmune lymphoproliferative syndrome type 4 Kras , Nras Autoinflammation, Antibody Deficiency, and Immune Dysregulation, PLCG2-Associated Plcg2 autosomal dominant cerebellar ataxia Prkcg autosomal dominant intellectual developmental disorder 48 Rarb autosomal dominant intellectual developmental disorder 5 Rxrb Autosomal Recessive Dyskeratosis Congenita Tp53 autosomal recessive dyskeratosis congenita 3 Tp53 autosomal recessive nonsyndromic deafness 97 Met autosomal recessive polycystic kidney disease Akt1 , Egfr , Pik3ca autosomal recessive spinocerebellar ataxia 14 Prkcg Autosomal Recessive Woolly Hair Rb1 B-Cell Chronic Lymphocytic Leukemia Braf , Hras , Kras , Nras , Plcg2 , Tp53 B-cell lymphoma Nras B-lymphoblastic leukemia/lymphoma Ccnd1 , Cdkn2a , Rb1 , Tp53 B-lymphoblastic leukemia/lymphoma with ETV6-RUNX1 Rb1 bacterial infectious disease Casp9 bacterial pneumonia Pten Balkan nephropathy Tp53 Bannayan-Riley-Ruvalcaba syndrome Egfr , Pik3ca , Pten Bardet-Biedl syndrome Akt3 Bardet-Biedl syndrome 16 Akt3 basal cell carcinoma Erbb2 , Pik3ca , Rb1 , Tp53 Behcet's disease Cdk6 bile duct adenoma Braf Bile Duct Neoplasms Egfr , Met biliary atresia Pten bipolar disorder Akt1 , Stk4 bladder carcinoma Hras bladder disease Rassf1 bladder squamous cell carcinoma Pten Bloom syndrome Map2k1 bone development disease Tp53 bone disease Cdkn2a Bone Marrow Failure Syndrome 5 Tp53 Bone Marrow Neoplasms Tp53 bone osteosarcoma Akt1 , Rb1 , Tp53 Bowen's Disease Ccnd1 brain disease Pten Brain Hypoxia-Ischemia Casp9 , Foxo3 Brain Injuries Akt1 , Bad brain ischemia Akt1 , Bad , Casp9 , Map2k1 , Mapk1 , Mapk3 , Prkca , Rb1 , Sos1 , Tp53 Brain Neoplasms Braf , Ccnd1 , Cdk4 , Cdkn2a , Egfr , Hras , Nras , Pik3ca , Pik3r1 , Pten , Tp53 Brain Stem Neoplasms Cdk4 , Cdk6 , Cdkn2a Brain-Lung-Thyroid Syndrome Sos2 breast adenocarcinoma Akt1 , Kras , Pik3ca , Tp53 breast angiosarcoma Pik3ca breast cancer Akt1 , Bad , Casp9 , Cdk4 , Cdkn2a , Egfr , Grb2 , Kras , Map2k1 , Map2k2 , Mapk1 , Met , Pik3ca , Plcg1 , Pten , Rassf1 , Tp53 Breast Cancer, Familial Akt1 , Erbb2 , Kras , Pik3ca , Pik3r3 , Pten , Tp53 breast carcinoma Cdk4 , Egfr , Met , Pik3ca , Plcg1 , Pten , Tp53 Breast Neoplasms Akt1 , Akt2 , Braf , Ccnd1 , Cdkn2a , E2f1 , Egf , Egfr , Erbb2 , Fhit , Hras , Kras , Pdpk1 , Pik3ca , Pik3r1 , Prkca , Pten , Raf1 , Rarb , Rassf1 , Rb1 , Rxrb , Tp53 bronchiolo-alveolar adenocarcinoma Ccnd1 , Egfr , Kras , Tp53 bronchopulmonary dysplasia Casp9 , Tp53 Brugada syndrome Sos1 Burkitt lymphoma Pik3r1 Burns Bad , Ccnd1 Cafe au lait Spots, Multiple Sos1 Calcification of Aortic Valve Akt1 , Tp53 calcinosis Erbb2 , Tp53 cancer Braf cannabis abuse Akt1 capillary hemangioma Akt3 Capillary Malformation-Arteriovenous Malformation 1 Kras Carcinogenesis Akt1 , Braf , Cdkn2a , Erbb2 , Kras , Met carcinoma Ccnd1 , Cdk4 , Egfr , Fhit , Hras , Kras , Met , Pik3r1 , Pten , Rb1 , Tp53 cardiac arrest Akt1 cardiofaciocutaneous syndrome Braf , Hras , Kras , Map2k1 , Map2k2 cardiofaciocutaneous syndrome 1 Braf , Kras , Map2k2 cardiofaciocutaneous syndrome 2 Kras cardiofaciocutaneous syndrome 3 Map2k1 cardiofaciocutaneous syndrome 4 Map2k2 Cardiomegaly Akt1 , Bad , Egfr , Mapk1 , Met , Prkca , Prkcb , Pten , Raf1 , Rb1 cardiomyopathy Akt1 , Braf , Kras , Mapk1 , Mapk3 , Sos1 , Tp53 Cardiotoxicity Tp53 Cardiovascular Abnormalities Map2k1 , Pik3ca , Pten carotid artery disease Tp53 Carotid Artery Injuries Foxo3 Carpenter Syndrome 2 Akt2 cataract Casp9 , Map2k1 , Pten , Tp53 Cecal Neoplasms Ccnd1 , Kras , Tp53 Central Nervous System Neoplasms Tp53 cerebellar ataxia Prkcg cerebral cavernous malformation Pten Cerebral Cavernous Malformation 4 Pik3ca Cerebral Hemorrhage Casp9 cerebral infarction Sos1 cerebral palsy Egfr cervical adenocarcinoma Tp53 cervical cancer Nras , Rassf1 , Sos1 , Tp53 cervical squamous cell carcinoma Tp53 cervix uteri carcinoma in situ Ccnd1 , Cdk4 , Cdkn2a , Pten , Rb1 Chanarin-Dorfman syndrome Hras Charcot-Marie-Tooth disease axonal type 2O Akt1 Charcot-Marie-Tooth disease dominant intermediate C Pik3r3 Charcot-Marie-Tooth disease type 2 Pik3cd Charcot-Marie-Tooth disease type 4 Foxo3 CHARGE syndrome Tp53 Chemical and Drug Induced Liver Injury Bad , E2f1 Chemotherapy-Induced Febrile Neutropenia Tp53 childhood hepatocellular carcinoma Met childhood pilocytic astrocytoma Mapk1 Chlamydia pneumonia Pik3cd Chlamydiaceae Infections Pten Chloracne Cdk6 , Egfr cholangiocarcinoma Braf , Cdkn2a , Egf , Egfr , Erbb2 , Kras , Met , Pten , Tp53 cholangitis Nras cholesteatoma of middle ear Tp53 chordoid glioma Prkca choroid plexus carcinoma Foxo3 , Tp53 choroid plexus papilloma Tp53 chromosome 13q14 deletion syndrome Rb1 chromosome 16p11.2 deletion syndrome, 593-kb Mapk3 chromosome 16p11.2 duplication syndrome Mapk3 Chromosome 17 Deletion Tp53 chromosome 1p36 deletion syndrome Casp9 , Pik3cd chromosome 22q11.2 deletion syndrome, distal Mapk1 Chromosome Aberrations Cdk6 Chromosome Deletion Cdkn2a Chronic Hepatitis Met Chronic Hepatitis C Ccnd1 chronic lymphocytic leukemia Braf , Hras , Kras , Nras , Tp53 chronic myelogenous leukemia, BCR-ABL1 positive Braf , Kras , Nras chronic myeloid leukemia Akt1 , Braf , Cdkn2a , E2f1 , Kras , Nras chronic myelomonocytic leukemia Nras chronic obstructive pulmonary disease Cdkn2a , Egfr , Foxo3 , Tp53 Chronic Pancreatitis Bad Circulating Neoplastic Cells Erbb2 CLAPO Syndrome Pik3ca clear cell adenocarcinoma Tp53 clear cell renal cell carcinoma Tp53 cleft palate Egf CLOVES syndrome Pik3ca , Pik3r1 cocaine abuse Mapk1 , Mapk3 cocaine dependence Mapk1 , Mapk3 Cocaine-Related Disorders Mapk1 , Mapk3 cognitive disorder Met Cognitive Dysfunction Tp53 coloboma Rarb colon adenocarcinoma Mapk1 , Mapk3 , Pik3ca , Pik3r3 colon adenoma Ccnd1 colon cancer Akt1 , Akt3 , Bad , Mapk1 , Pik3r2 , Pik3r5 , Plcg1 , Pten colon carcinoma Akt1 , Braf , Nras , Pik3ca , Pten , Tp53 Colon Diverticulum Tp53 Colonic Neoplasms Braf , Ccnd1 , Cdkn2a , Egfr , Erbb2 , Kras , Pik3r1 , Pik3r2 , Tp53 colorectal adenocarcinoma Braf , Mapk1 , Mapk3 , Raf1 , Tp53 colorectal adenoma Raf1 colorectal cancer Akt1 , Akt2 , Braf , Casp9 , Ccnd1 , Egfr , Erbb2 , Kras , Map2k1 , Map2k2 , Met , Nras , Pik3ca , Pik3cb , Pik3cd , Pik3r1 , Plcg1 , Pten , Tp53 colorectal carcinoma Pik3ca , Tp53 Colorectal Neoplasms Akt1 , Akt2 , Braf , Ccnd1 , Cdkn2a , Egfr , Erbb2 , Hras , Kras , Map2k1 , Mapk1 , Mapk3 , Met , Nras , Pik3ca , Pik3r1 , Pik3r3 , Prkcb , Pten , Raf1 , Tp53 combined oxidative phosphorylation deficiency 11 Tp53 common variable immunodeficiency Tp53 complex cortical dysplasia with other brain malformations Akt3 , Hras , Pik3ca congenital diaphragmatic hernia Met congenital fibrosarcoma Tp53 congenital fibrosis of the extraocular muscles Hras congenital heart disease Mapk1 , Rxra , Sos1 Congenital Hypomyelinating Neuropathy Pten congenital hypoplastic anemia Akt2 congenital merosin-deficient muscular dystrophy 1A Tp53 congenital myasthenic syndrome 2A Tp53 Congenital Myopathy with Excess of Muscle Spindles Hras congestive heart failure Erbb2 , Map2k1 , Pdpk1 , Prkca , Prkcb , Tp53 Copper-Overload Cirrhosis Met coronary artery disease Akt1 , Tp53 coronary restenosis Mapk1 , Tp53 Coronavirus infectious disease Egfr , Mapk1 , Mapk3 coronin-1A deficiency Mapk3 Costello syndrome Braf , Hras , Kras , Map2k1 , Map2k2 COVID-19 Akt3 , E2f1 Cowden syndrome Pik3ca , Pten Cowden syndrome 4 Pten Cowden syndrome 5 Pik3ca Cowden syndrome 6 Akt1 Craniofacial Abnormalities Egfr , Pten , Tp53 craniopharyngioma Braf craniosynostosis Akt2 Cryopyrin-Associated Periodic Syndromes Plcg2 Currarino syndrome Pik3ca cutaneous leishmaniasis Pik3cd cutaneous Paget's disease Pik3ca Cystic, Mucinous, and Serous Neoplasms Met Dandy-Walker syndrome Braf delta beta-thalassemia Hras dementia E2f1 , Egfr demyelinating disease Mapk1 , Tp53 depressive disorder Mapk1 dermatitis Egf , Egfr dermoid cyst of ovary Foxo3 developmental and epileptic encephalopathy Hras , Pik3r3 , Rassf1 developmental and epileptic encephalopathy 14 Rxra developmental and epileptic encephalopathy 54 Akt3 developmental coordination disorder Pten Developmental Disabilities Akt3 , Braf , Mapk3 , Pik3ca , Pik3r2 , Pten Developmental Disease Sos2 Diabetes Complications Casp9 diabetes mellitus Egfr , Pten Diabetic Cardiomyopathies Akt1 , Bad , Casp9 , Erbb2 , Mapk1 , Mapk3 , Pik3ca Diabetic Cystopathy Bad Diabetic Nephropathies Casp9 , Egf , Mapk1 , Prkcb , Pten , Tp53 diabetic neuropathy Bad diabetic retinopathy Akt1 , Casp9 , Egfr , Hras , Pten Diamond-Blackfan anemia Akt2 , Tp53 diaphragmatic eventration Pik3ca Diaphragmatic Hernia Rxra diffuse large B-cell lymphoma Braf , Cdkn2a , Pdpk1 , Pik3ca , Pik3cb , Pik3cd , Pten , Tp53 diffuse pediatric-type high-grade glioma, H3-wildtype and IDH-wildtype Cdk4 , Tp53 DiGeorge syndrome Mapk1 , Tp53 dilated cardiomyopathy Braf , Casp9 , Egfr , Erbb2 , Kras , Prkca , Prkcb , Raf1 , Sos1 , Tp53 dilated cardiomyopathy 1B Raf1 dilated cardiomyopathy 1NN Raf1 Dilated Cardiomyopathy with Left Ventricular Noncompaction Raf1 disease of cellular proliferation Braf , Cdkn2a , E2f1 , Erbb2 , Hras , Kras , Map2k1 , Met , Nras , Pik3ca , Pik3r1 , Pten , Rb1 , Tp53 Disease Progression Ccnd1 , Egf , Egfr , Erbb2 , Hras , Kras , Pik3ca , Pten , Rxrb , Tp53 distal arthrogryposis type 1 Met Distal Arthrogryposis Type 11 Met distal arthrogryposis type 1A Met Down syndrome Pik3ca Drug-induced Neutropenia Tp53 Drug-Related Side Effects and Adverse Reactions Tp53 Ductal Carcinoma Pten , Tp53 ductal carcinoma in situ Ccnd1 , Erbb2 , Prkca , Rb1 , Tp53 Dwarfism Mapk1 , Sos1 , Tp53 dysembryoplastic neuroepithelial tumor Braf dyskeratosis congenita Tp53 dysplastic nevus syndrome Braf , Cdkn2a early infantile epileptic encephalopathy Hras , Pik3r3 , Rassf1 Edema Casp9 Ehlers-Danlos syndrome classic type 1 Rxra Embryo Loss Foxo3 embryonal carcinoma Rb1 Emphysema Tp53 Encephalocraniocutaneous Lipomatosis Kras endometrial adenocarcinoma Foxo3 , Kras , Mapk1 , Mapk3 , Pik3r1 , Pten , Tp53 endometrial cancer Akt2 , Bad , Cdkn2a , Egf , Pik3cb , Pik3cd , Pik3r1 , Pten , Tp53 endometrial carcinoma Braf , Egfr , Grb2 , Hras , Kras , Mapk1 , Mapk3 , Pik3ca , Pik3r1 , Prkca , Pten , Rassf1 , Tp53 endometrial hyperplasia Kras , Pten Endometrial Hyperplasia without Atypia Kras Endometrial Intraepithelial Neoplasia Cdkn2a , Kras , Pten , Tp53 Endometrial Neoplasms Ccnd1 , Cdk4 , Cdkn2a , Egf , Erbb2 , Fhit , Kras , Mapk3 , Met , Pik3ca , Pik3cb , Pten , Rb1 , Tp53 Endometrioid Carcinomas Akt1 , Egfr , Erbb2 , Kras , Mapk1 , Mapk3 , Pik3r2 , Pten endometriosis Egfr , Kras , Rarb endometritis Casp9 Endotoxemia Akt1 , Casp9 epidermal nevus Hras , Kras , Nras , Pik3ca epilepsy Akt1 , Pdpk1 , Pik3r2 , Pten episodic kinesigenic dyskinesia 1 Mapk3 Epstein-Barr virus infectious disease Pten esophageal cancer Akt1 , Pik3ca , Pten , Tp53 esophageal carcinoma Cdkn2a , Egfr , Erbb2 , Hras , Kras , Pik3ca , Tp53 Esophageal Neoplasms Ccnd1 , Cdkn2a , Egfr , Erbb2 , Met , Tp53 esophagus adenocarcinoma Cdkn2a , Pik3ca , Tp53 esophagus squamous cell carcinoma Akt1 , Ccnd1 , Cdkn2a , Egfr , Rb1 , Tp53 estrogen-receptor positive breast cancer Pik3ca Ewing sarcoma Cdk4 Experimental Arthritis Akt1 , Cdkn2a , Egfr , Nras , Pik3cd Experimental Autoimmune Encephalomyelitis Casp9 , Foxo3 Experimental Autoimmune Myocarditis Casp9 Experimental Colitis Akt1 , Mapk1 , Mapk3 Experimental Diabetes Mellitus Akt1 , Bad , Casp9 , Egfr , Foxo3 , Hras , Mapk1 , Mapk3 , Met , Pik3r1 , Plcg1 , Prkca , Prkcb , Tp53 Experimental Leukemia Cdkn2a , Nras Experimental Liver Cirrhosis Cdk4 , Egf , Mapk1 , Mapk3 , Met , Prkcb , Rxra Experimental Liver Neoplasms Ccnd1 , Hras , Kras , Map2k1 , Mapk1 , Mapk3 , Met , Nras , Rb1 , Rxra , Tgfa , Tp53 Experimental Mammary Neoplasms Akt1 , Akt2 , Bad , Casp9 , Ccnd1 , Cdk4 , Cdk6 , Cdkn2a , Egfr , Erbb2 , Hras , Mapk1 , Mapk3 , Pik3r1 , Pten , Rb1 , Rxra , Tgfa , Tp53 Experimental Melanoma Braf , Pten Experimental Neoplasms Braf , Ccnd1 , Cdk4 , Erbb2 , Hras , Kras , Nras , Rb1 , Tp53 Experimental Radiation Injuries Casp9 , Mapk1 , Mapk3 Experimental Sarcoma Kras , Nras extrahepatic cholestasis Mapk1 , Mapk3 Eye Neoplasms Tp53 Facial Asymmetry Pik3ca familial adenomatous polyposis Akt1 , Egfr , Plcg1 familial adult myoclonic epilepsy 5 Rassf5 familial cold autoinflammatory syndrome Plcg2 familial cold autoinflammatory syndrome 2 Prkcg familial cold autoinflammatory syndrome 3 Plcg2 familial hypertrophic cardiomyopathy Raf1 , Sos1 familial melanoma Braf , Cdk4 , Cdkn2a , Tp53 familial meningioma Pten familial multiple nevi flammei Pik3ca familial partial lipodystrophy Akt2 Familial Prostate Cancer Map2k1 familial retinoblastoma Rb1 Familial Visceral Neuropathy 2, Autosomal Recessive Erbb2 Female Infertility Mapk1 , Mapk3 , Tp53 fetal alcohol spectrum disorder Pten Fetal Diseases Prkcb Fever Plcg2 fibrillary astrocytoma Kras Fibrosis Akt1 Fluoride Poisoning Cdkn2a focal epilepsy Nras , Plcg1 , Stk4 focal segmental glomerulosclerosis Casp9 focal segmental glomerulosclerosis 5 Akt1 Follicular Thyroid Cancer Braf , Hras , Nras , Pten Funnel Chest Tp53 gallbladder cancer Braf , Egf , Kras , Pik3ca , Tp53 gallbladder carcinoma Erbb2 , Pik3ca , Tgfa , Tp53 Gallbladder Neoplasms Egfr , Erbb2 , Kras , Met , Tp53 ganglioglioma Braf gastric adenocarcinoma Akt1 , Braf , Cdkn2a , Erbb2 , Hras , Kras , Map2k1 , Map2k2 , Nras , Pik3ca , Pten , Raf1 , Tp53 gastric ulcer Bad , Egf , Mapk1 , Mapk3 gastrinoma Met gastroenteritis Tgfa Gastrointestinal Neoplasms Erbb2 , Pik3ca , Pten gastrointestinal stromal tumor Akt3 , Braf , Kras , Rassf5 , Rb1 , Rxrg Generalized Epilepsy Pten genetic disease Akt1 , Akt2 , Akt3 , Bad , Braf , Casp9 , Ccnd1 , Cdk6 , Cdkn2a , E2f1 , E2f3 , Egf , Egfr , Erbb2 , Fhit , Foxo3 , Grb2 , Hras , Kras , Map2k1 , Map2k2 , Mapk1 , Mapk3 , Met , Nras , Pdpk1 , Pik3ca , Pik3cb , Pik3cd , Pik3r1 , Pik3r2 , Pik3r3 , Pik3r5 , Plcg1 , Plcg2 , Prkca , Prkcb , Prkcg , Pten , Raf1 , Rarb , Rassf1 , Rassf5 , Rb1 , Rxra , Rxrb , Rxrg , Sos1 , Sos2 , Stk4 , Tgfa Genetic Predisposition to Disease Egf , Tgfa Genetic Translocation Cdkn2a Genomic Instability Ccnd1 Germ Cell and Embryonal Neoplasms Cdk4 , Cdkn2a , Erbb2 , Fhit germinoma Braf , Cdkn2a gestational diabetes Met Gilles de la Tourette syndrome Met gingival fibromatosis Sos1 Gingival Fibromatosis 1 Sos1 glaucoma Bad , Cdkn2a glioblastoma Akt1 , Akt3 , Braf , Cdk4 , Cdk6 , Cdkn2a , E2f1 , Egf , Egfr , Hras , Kras , Map2k2 , Met , Nras , Pik3ca , Pik3cb , Pik3cd , Pik3r1 , Pten , Rb1 , Tp53 glioblastoma proneural subtype Rb1 Gliosis Mapk1 , Mapk3 glomerulonephritis Mapk1 glucose intolerance Pik3r1 glycogen storage disease II Pik3ca Glycogen Storage Disease XII Mapk3 graft-versus-host disease Tp53 granulosa cell tumor Cdkn2a , Fhit Graves' disease Tp53 Growth Disorders Pik3ca Habitual Abortions Foxo3 hairy cell leukemia Map2k1 head and neck cancer Egfr , Pik3ca , Tp53 Head and Neck Neoplasms Egfr , Mapk1 , Mapk3 , Pik3ca , Rarb , Tgfa , Tp53 head and neck squamous cell carcinoma Akt1 , Braf , Ccnd1 , Cdk4 , Cdk6 , Cdkn2a , Egfr , Erbb2 , Fhit , Hras , Map2k1 , Map2k2 , Mapk1 , Pik3ca , Pten , Rb1 , Tp53 heart disease Casp9 Helicobacter Infections Egf , Egfr , Erbb2 , Pten HELLP syndrome Erbb2 hemangioma Foxo3 , Pten hemangiopericytoma Pten hematologic cancer Tp53 Hemimegalencephaly Pik3ca , Pten Hemorrhagic Shock Bad , Mapk1 hepatitis B Pten hepatoblastoma Met , Tgfa , Tp53 hepatocellular adenoma Hras , Kras , Pten hepatocellular carcinoma Akt1 , Akt2 , Braf , Casp9 , Ccnd1 , Cdk4 , Cdkn2a , E2f1 , Egf , Egfr , Fhit , Hras , Kras , Map2k2 , Mapk1 , Met , Nras , Pik3ca , Pik3cb , Pik3r1 , Pik3r2 , Pik3r3 , Plcg1 , Pten , Rb1 , Rxra , Tgfa , Tp53 Hepatomegaly Akt1 , Met , Pten Hereditary Adrenocortical Carcinoma Tp53 hereditary ataxia Prkcg hereditary breast ovarian cancer syndrome Egfr , Kras , Plcg1 , Pten , Tp53 hereditary diffuse gastric cancer Kras Hereditary Neoplastic Syndromes Akt1 , Cdk4 , Cdkn2a , Egfr , Hras , Met , Pik3ca , Pten , Rb1 , Tp53 hereditary nonpolyposis colorectal cancer type 4 Rb1 Hereditary Pancreatitis Casp9 hereditary renal cell carcinoma Egf hereditary spastic paraplegia 47 Nras high grade glioma Akt2 , Bad , Braf , Ccnd1 , Cdk4 , Cdk6 , Cdkn2a , E2f1 , E2f3 , Egf , Egfr , Erbb2 , Hras , Kras , Map2k1 , Mapk3 , Met , Pik3ca , Pik3r3 , Prkca , Pten , Raf1 , Rassf1 , Tp53 Hirschsprung's disease Erbb2 Hodgkin's lymphoma Kras , Mapk1 , Met Huntington's disease Casp9 , E2f1 , Egfr Hydrops Fetalis Hras , Kras Hyperalgesia Mapk1 , Mapk3 , Prkcb , Prkcg , Raf1 Hypercholesterolemia Casp9 hyperglycemia Prkcb hyperinsulinism Pten Hyperoxia Cdk4 hyperparathyroidism Ccnd1 Hyperplasia Akt1 , Ccnd1 , Cdkn2a , Egfr , Pik3ca Hypertelorism Pik3ca hypertension Akt1 , Akt2 , Bad , Casp9 , Cdkn2a , Egfr , Kras , Pik3r1 , Prkca , Prkcb , Pten , Tp53 Hypertriglyceridemia Akt1 , Met hypertrophic cardiomyopathy Braf , Map2k2 , Raf1 , Rassf1 , Sos1 , Tp53 hypertrophic cardiomyopathy 1 Raf1 , Sos1 hypertrophic cardiomyopathy 25 Erbb2 hypertrophic cardiomyopathy 4 Braf Hypertrophy Mapk1 , Mapk3 , Pten hypoglycemia Akt2 , Pik3r1 Hypogonadism and Testicular Atrophy Tp53 hypoinsulinemic hypoglycemia with hemihypertrophy Akt2 Hypokalemic Periodic Paralysis, Type 1 Rassf5 Hypophosphatemic Rickets Hras hypothyroidism Bad hypotrichosis 8 Rb1 idiopathic generalized epilepsy Pdpk1 idiopathic pulmonary fibrosis Akt1 Immediate Hypersensitivity Egfr IMMUNE DYSREGULATION, AUTOIMMUNITY, AND AUTOINFLAMMATION Plcg1 immune system disease Prkcb immunodeficiency 14 Pik3cd immunodeficiency 36 Pik3r1 immunodeficiency 39 Hras impotence Akt1 , Bad , Ccnd1 in situ carcinoma Akt1 , Cdkn2a , Pten , Rb1 inclusion body myopathy with Paget disease of bone and frontotemporal dementia Egf Inflammation Akt1 , Egfr , Tgfa inflammatory bowel disease Plcg2 , Rassf1 Insulin Resistance Akt2 , Bad , Egfr , Mapk3 , Pik3r1 , Prkcb , Pten , Rb1 insulinoma Tgfa intellectual disability Bad , Braf , Ccnd1 , Hras , Kras , Mapk1 , Met , Pik3r2 , Prkcg , Pten , Rb1 , Sos1 intermediate coronary syndrome Akt1 , Pten intermittent claudication Bad interstitial cystitis Erbb2 Intervertebral Disc Displacement Akt1 Intestinal Neoplasms Prkca Intestinal Polyps Ccnd1 intestinal pseudo-obstruction Pten , Rb1 Intestinal Reperfusion Injury Casp9 Intimal Hyperplasia Foxo3 , Mapk1 , Pten intrahepatic cholangiocarcinoma Braf , Kras , Pten invasive ductal carcinoma Akt1 , Cdkn2a , Erbb2 , Fhit , Met invasive lobular carcinoma Ccnd1 , Erbb2 ischemia Akt2 , Prkca , Raf1 , Tp53 islet cell tumor Cdk4 Joubert syndrome 3 Sos1 juvenile myelomonocytic leukemia Kras , Nras juvenile polyposis syndrome Pten juvenile rheumatoid arthritis Egf , Kras , Mapk1 Kaposi's sarcoma Tp53 keratoacanthoma Pik3ca keratosis Tp53 kidney disease Casp9 , Egf , Mapk1 Kidney Neoplasms Akt1 , Braf , Ccnd1 , Cdkn2a , Erbb2 , Kras , Mapk1 , Mapk3 , Met , Raf1 , Rassf1 , Rassf5 Klatskin's tumor Kras Kleefstra syndrome 1 Rxra Klippel-Trenaunay syndrome Pik3ca Knee Osteoarthritis Tgfa L-2-hydroxyglutaric aciduria Sos2 Langerhans-cell histiocytosis Braf Language Development Disorders Pten large cell carcinoma Met , Prkca large congenital melanocytic nevus Hras , Nras Laryngeal Neoplasms Egfr laryngeal squamous cell carcinoma Egfr , Erbb2 , Plcg1 Leber congenital amaurosis Hras , Pik3ca Leber hereditary optic neuropathy Tp53 Left Ventricular Hypertrophy Egfr , Foxo3 , Mapk1 , Prkca , Prkcb , Pten Leigh disease Rxra leiomyoma Egfr , Pten leiomyosarcoma Cdkn2a leukemia Bad , Ccnd1 , Cdkn2a , Nras , Pten , Tp53 leukocyte adhesion deficiency 3 Bad Leukocytosis Kras , Nras Leydig cell tumor Cdk4 Li-Fraumeni syndrome Tp53 Li-Fraumeni syndrome 1 Tp53 Li-Fraumeni-Like Syndrome Tp53 linear nevus sebaceous syndrome Hras , Kras , Nras Lip Neoplasms Tp53 lipoma Pten lipomatosis Pten liposarcoma Cdk4 listeriosis Pik3cd liver benign neoplasm Met liver cancer Ccnd1 , Hras , Pik3ca liver carcinoma Hras , Nras liver cirrhosis Casp9 , Egfr , Foxo3 , Hras , Pten Liver Metastasis Braf , Nras Liver Neoplasms Braf , Ccnd1 , Cdkn2a , Fhit , Hras , Kras , Raf1 , Tgfa , Tp53 Liver Reperfusion Injury Bad , Casp9 long QT syndrome E2f1 , Pten low tension glaucoma Tp53 lung adenocarcinoma Akt1 , Braf , Ccnd1 , Cdk4 , Cdk6 , Cdkn2a , E2f1 , E2f3 , Egf , Egfr , Erbb2 , Foxo3 , Hras , Kras , Map2k1 , Nras , Pik3ca , Pik3r1 , Plcg1 , Pten , Raf1 , Rarb , Rb1 , Sos1 , Tp53 lung cancer Braf , Egfr , Erbb2 , Kras , Pik3ca , Plcg1 lung carcinoma Braf , Egfr , Kras , Met , Pik3ca lung disease Egfr Lung Injury Tgfa Lung Neoplasms Akt1 , Braf , Ccnd1 , Cdkn2a , Egfr , Erbb2 , Fhit , Hras , Kras , Mapk1 , Mapk3 , Met , Pik3ca , Pten , Raf1 , Rarb , Rassf1 , Tp53 lung non-small cell carcinoma Akt1 , Akt2 , Akt3 , Bad , Braf , Casp9 , Ccnd1 , Cdk4 , Cdk6 , Cdkn2a , E2f1 , E2f3 , Egf , Egfr , Erbb2 , Fhit , Foxo3 , Hras , Kras , Map2k1 , Map2k2 , Met , Nras , Pdpk1 , Pik3ca , Plcg1 , Prkcb , Pten , Raf1 , Rarb , Rassf1 , Rassf5 , Rb1 , Rxrg , Tgfa , Tp53 lung oat cell carcinoma Akt1 , Pik3ca , Pten , Rb1 , Tp53 Lung Reperfusion Injury Kras lung sarcomatoid carcinoma Kras , Tp53 lung small cell carcinoma Akt1 , Ccnd1 , E2f1 , Egfr , Pik3ca , Pik3r1 , Pten , Rarb , Rb1 , Tp53 lung squamous cell carcinoma Akt1 , Braf , Ccnd1 , Cdk4 , Cdkn2a , E2f1 , E2f3 , Egfr , Hras , Kras , Map2k1 , Met , Pik3ca , Pten , Tp53 lupus nephritis Egf Lymphangiectasis Raf1 lymphangioma Braf Lymphatic Metastasis Braf , Casp9 , Cdkn2a , Egfr , Erbb2 , Met , Pdpk1 , Pik3ca , Pik3cb , Pten , Tp53 lymphedema Met lymphoblastic lymphoma Foxo3 lymphoma Cdkn2a , Kras , Nras , Pten , Tp53 Lynch syndrome Kras , Sos1 Macrocephaly Akt3 , Pik3ca , Pten Macrocephaly Mesodermal Hamartoma Spectrum Akt1 macrocephaly-autism syndrome Pten macular degeneration Bad Maffucci syndrome Cdkn2a major depressive disorder Pten male infertility Tp53 malignant astrocytoma Akt1 , Akt2 , Akt3 , Braf , Ccnd1 , Kras , Met , Tp53 malignant mesothelioma Cdk6 , Cdkn2a , Egfr , Met , Raf1 , Rxra , Tp53 mammary Paget's disease Erbb2 Mandibular Neoplasms Braf mantle cell lymphoma Ccnd1 , Pik3cd maple syrup urine disease Akt2 marginal zone lymphoma Tp53 maturity-onset diabetes of the young type 1 Akt2 median neuropathy Egfr , Erbb2 medulloblastoma Ccnd1 , Cdk6 , Erbb2 , Foxo3 , Nras , Pik3ca , Rassf1 , Rb1 , Tp53 megacolon Akt3 , Fhit Megalencephaly - Cutis Marmorata Telangiectatica Congenita Akt3 , Pik3ca , Pik3r1 , Pik3r2 Megalencephaly-Polymicrogyria-Polydactyly-Hydrocephalus Syndrome Akt3 , Pik3ca , Pik3r2 Megalencephaly-Polymicrogyria-Polydactyly-Hydrocephalus Syndrome 1 Pik3r2 Megalencephaly-Polymicrogyria-Polydactyly-Hydrocephalus Syndrome 2 Akt3 Megalodactyly Pik3ca melanoma Akt1 , Braf , Cdk4 , Cdkn2a , E2f1 , Erbb2 , Hras , Kras , Map2k1 , Map2k2 , Mapk1 , Met , Nras , Pik3ca , Pik3r1 , Pten , Raf1 , Tp53 melanoma and neural system tumor syndrome Cdkn2a Melanoma-Pancreatic Cancer Syndrome Cdkn2a melorheostosis Map2k1 Memory Disorders Akt1 , Mapk1 , Mapk3 Meningeal Neoplasms Akt1 meninges sarcoma Tp53 meningioma Akt1 , Pten Mesothelioma Cdkn2a , Egfr , Fhit , Rassf1 , Tp53 metabolic dysfunction-associated steatotic liver disease Met , Prkca , Pten Metabolic Syndrome Map2k2 Metaplasia Egf Micronuclei, Chromosome-Defective Cdkn2a , Rassf1 microphthalmia Rarb Microsatellite Instability Braf middle cerebral artery infarction Bad , Casp9 , Mapk1 , Mapk3 , Prkcg migraine Map2k2 mismatch repair cancer syndrome Tp53 mitochondrial DNA depletion syndrome 5 Rb1 mitochondrial metabolism disease Tp53 morphine dependence Mapk1 , Mapk3 Mouth Neoplasms Cdkn2a , Erbb2 , Hras , Kras , Nras , Pik3ca , Pten , Tp53 mucositis Tgfa Muir-Torre syndrome Fhit multicentric Castleman disease Map2k2 Multiple Abnormalities Kras , Pten multiple myeloma Braf , Ccnd1 , Cdk4 , Cdkn2a , Hras , Kras , Met , Nras , Pik3ca , Pik3r2 , Rxra , Tp53 Multiple Organ Failure Mapk1 Muscle Hypotonia Sos1 muscular atrophy Akt1 , Kras muscular dystrophy Pten myelodysplastic syndrome Cdkn2a , Erbb2 , Hras , Kras , Nras , Tp53 myelofibrosis Tp53 myeloid leukemia associated with Down Syndrome Nras , Tp53 myeloid neoplasm Kras , Nras , Tp53 myeloid sarcoma Kras myeloproliferative neoplasm Pten myocardial infarction Akt1 , Erbb2 , Mapk1 , Tp53 Myocardial Ischemia Ccnd1 , Erbb2 , Rxrg Myocardial Reperfusion Injury Akt1 , Akt2 , Bad , Casp9 , Mapk1 , Mapk3 , Prkcb , Pten , Tp53 myoepithelioma Braf nasal cavity cancer Tp53 Nasal Polyps Met Nasopharyngeal Neoplasms Ccnd1 , Egfr , Hras , Nras , Pik3ca , Tp53 nasopharynx carcinoma Tp53 NATURAL KILLER CELL ENTEROPATHY Pik3cb Nematode Infections Pik3r1 Neointima Akt1 Neonatal Hypoglycemia, Simulating Foetopathia Diabetica Akt2 Neonatal Inflammatory Skin and Bowel Disease 2 Egfr Neoplasm Invasiveness Cdkn2a , Egf , Egfr , Hras , Mapk1 , Mapk3 , Met , Pten Neoplasm Metastasis Akt2 , Braf , Ccnd1 , Cdkn2a , E2f1 , Egf , Egfr , Erbb2 , Kras , Mapk1 , Mapk3 , Met , Pik3ca , Pik3r3 , Pten , Rb1 , Rxra , Tp53 Neoplasm Recurrence, Local Egfr , Erbb2 Neoplastic Cell Transformation Ccnd1 , Egf , Hras , Kras , Pten , Tp53 Neoplastic Processes Hras nephroblastoma Braf , Cdk4 , Cdkn2a , Erbb2 , Tp53 nephrogenic diabetes insipidus Prkca Nerve Degeneration Mapk1 , Mapk3 , Tgfa Nerve Sheath Neoplasms Cdkn2a , Met , Pten , Tp53 nervous system disease Casp9 , Tp53 Nervous System Malformations Pten neurilemmoma Cdkn2a neuroblastoma Cdkn2a , Hras , Kras , Met , Nras , Pik3ca , Rassf1 , Rassf5 , Tp53 Neurocutaneous Melanosis Nras NEURODEVELOPMENTAL DISORDER WITH MICROCEPHALY, ARTHROGRYPOSIS, AND STRUCTURAL BRAIN ANOMALIES Pik3ca Neurodevelopmental Disorders Akt3 , Mapk3 , Nras , Pik3cd , Pik3r1 , Pten , Sos1 neuroendocrine tumor Braf neurofibromatosis-Noonan syndrome Map2k2 neurofibrosarcoma Nras Neurogenic Inflammation Rxra neuronal ceroid lipofuscinosis Hras , Map2k1 neuronal ceroid lipofuscinosis 6A Mapk1 , Mapk3 Nevus Braf newborn respiratory distress syndrome Braf NGLY1-deficiency Rarb non-functioning pancreatic endocrine tumor Cdk4 , Cdk6 non-Hodgkin lymphoma Braf , Ccnd1 , Map2k1 , Nras , Pik3ca , Tp53 Nonseminomatous Germ Cell Tumor Braf nonsyndromic deafness Met , Tgfa Noonan Like Syndrome Sos1 Noonan syndrome Braf , Hras , Kras , Map2k1 , Map2k2 , Nras , Raf1 , Sos1 , Sos2 Noonan syndrome 1 Braf , Hras , Kras , Map2k1 , Map2k2 , Nras , Raf1 , Sos1 , Sos2 Noonan syndrome 13 Mapk1 Noonan syndrome 3 Hras , Kras , Raf1 , Sos1 Noonan syndrome 4 Sos1 Noonan syndrome 5 Raf1 Noonan syndrome 6 Nras Noonan syndrome 7 Braf Noonan syndrome 8 Pik3ca Noonan syndrome 9 Sos2 Noonan syndrome with multiple lentigines Braf , Map2k2 , Pten , Raf1 Noonan syndrome with multiple lentigines 2 Raf1 Noonan syndrome with multiple lentigines 3 Braf NSAID-Enteropathy Casp9 Nuchal Bleb, Familial Sos1 obesity Akt1 , Akt2 , Bad , Cdk4 , Cdkn2a , Egfr , Foxo3 , Mapk1 , Mapk3 , Pik3r1 , Prkcb , Rb1 , Rxra obsessive-compulsive disorder Hras occupational asthma Prkca oculoectodermal syndrome Kras Odontogenic Myxoma Tp53 oligodendroglioma Egf , Rb1 Oral Lichen Planus Tp53 oral mucosa leukoplakia Tp53 oral squamous cell carcinoma Egfr , Erbb2 , Met , Plcg1 , Pten , Tp53 oral submucous fibrosis Tp53 Oropharyngeal Neoplasms Tp53 osteoarthritis Bad , Casp9 , Cdkn2a , Pik3r1 Osteoarthritis, Experimental Mapk3 osteochondrodysplasia Pten Osteofibrous Dysplasia Met osteosarcoma Akt1 , Cdkn2a , E2f1 , Egfr , Met , Rb1 , Tp53 ovarian cancer Akt1 , Cdk4 , Cdkn2a , Egf , Egfr , Erbb2 , Hras , Met , Pik3ca , Prkca , Pten , Rassf1 , Rb1 , Tp53 ovarian carcinoma Akt2 , Ccnd1 , Erbb2 , Map2k1 , Prkca , Raf1 , Tp53 Ovarian Neoplasms Akt1 , Akt2 , Braf , Cdk4 , Cdkn2a , Egfr , Erbb2 , Fhit , Hras , Kras , Map2k1 , Map2k2 , Mapk1 , Mapk3 , Met , Pik3ca , Pik3r1 , Pten , Rassf1 , Rb1 , Tp53 ovarian serous carcinoma Pten ovarian serous cystadenocarcinoma Braf , Erbb2 , Hras , Nras , Pik3ca , Tp53 ovarian small cell carcinoma Tp53 ovary adenocarcinoma Erbb2 ovary serous adenocarcinoma Kras , Tp53 Pain Akt1 pancreatic acinar cell adenocarcinoma Braf pancreatic adenocarcinoma Akt3 , Ccnd1 , Cdk4 , Cdk6 , Cdkn2a , E2f1 , Erbb2 , Hras , Kras , Map2k1 , Map2k2 , Mapk1 , Pik3ca , Raf1 , Rxra , Tp53 pancreatic cancer Akt1 , Akt2 , Bad , Braf , Casp9 , Ccnd1 , Cdk4 , Cdkn2a , E2f1 , Egf , Egfr , Kras , Met , Pten , Rb1 , Rxra , Tgfa , Tp53 pancreatic carcinoma Cdkn2a , Kras , Raf1 , Tp53 pancreatic ductal adenocarcinoma Ccnd1 , Cdk4 , E2f1 , Pik3r2 pancreatic ductal carcinoma Ccnd1 , Cdk6 , Cdkn2a , Egf , Kras , Mapk3 , Met , Rxra , Tp53 pancreatic intraductal papillary-mucinous neoplasm Akt1 , Tp53 Pancreatic Intraepithelial Neoplasia Ccnd1 pancreatic mucinous cystadenoma Kras pancreatic solid pseudopapillary carcinoma Ccnd1 pancreatitis Egf , Map2k1 , Met pancytopenia Tp53 papillary renal cell carcinoma Braf , Cdkn2a , Erbb2 , Hras , Met , Pik3ca , Tp53 papillary tumor of the pineal region Pten papilloma Ccnd1 , Egfr , Erbb2 , Hras , Tgfa papillomavirus infectious disease Cdkn2a , Egfr , Pten paranasal sinus benign neoplasm Tp53 paraplegia Akt1 Paraproteinemias Braf , Hras , Kras , Nras , Pik3ca , Tp53 Parasitic Liver Diseases Kras , Tp53 parathyroid adenoma Hras parathyroid carcinoma Akt3 , Rassf5 , Rxrg Parkinson's disease Akt1 , Casp9 , Egf , Egfr , Erbb2 , Mapk1 , Mapk3 Parkinsonism Bad , Casp9 , Egfr Pediatric Adrenocortical Carcinoma Tp53 penile benign neoplasm Ccnd1 Penile Neoplasms Hras , Kras , Pik3ca , Tp53 peptic esophagitis Egfr perinatal necrotizing enterocolitis Egf periodontitis Met peripheral nervous system disease Casp9 , Cdkn2a peripheral nervous system neoplasm Pten persistent fetal circulation syndrome Pten persistent hyperplastic primary vitreous Cdkn2a PHACE Association Braf phobic disorder Hras Phyllodes Tumor Egfr , Rb1 , Tp53 Pigmented Nevus Hras , Nras pilocytic astrocytoma Kras Pituitary Neoplasms Prkca , Rb1 Pituitary Stalk Interruption Syndrome Raf1 plasma cell neoplasm Braf , Hras , Kras , Nras , Pik3ca , Tp53 plasmacytoma Cdkn2a pleomorphic xanthoastrocytoma Braf , Cdk6 , Egfr , Met , Tp53 Pneumococcal Pneumonia Mapk1 , Mapk3 , Pten pneumonia Egf Poisoning Cdkn2a polycystic kidney disease Pik3ca , Plcg1 polycystic ovary syndrome Akt2 polydactyly Pten polymicrogyria Akt3 Polyploidy Tp53 Polyuria Prkca portal hypertension Mapk1 , Pik3r1 , Pten post-traumatic stress disorder Casp9 Postoperative Cognitive Dysfunction Mapk1 , Mapk3 pre-eclampsia Erbb2 pre-malignant neoplasm Akt1 , Casp9 , Ccnd1 , Cdkn2a , Egf , Hras , Tp53 Premature Aging Cdkn2a , Tp53 Premature Birth Braf primary autosomal recessive microcephaly 12 Cdk6 primary biliary cholangitis Met primary cerebellar degeneration Prkcg primary ciliary dyskinesia Rassf1 primary coenzyme Q10 deficiency 7 Rxra primary cutaneous T-cell non-Hodgkin lymphoma Braf , Cdkn2a , Plcg1 , Tp53 primary hypomagnesemia Egf primary immunodeficiency disease Pik3cd , Pik3r1 , Stk4 primary open angle glaucoma Tp53 prostate adenocarcinoma Akt1 , Akt2 , Akt3 , Braf , Cdkn2a , Erbb2 , Grb2 , Hras , Map2k1 , Mapk1 , Pdpk1 , Pik3ca , Pik3cb , Pten , Raf1 , Tp53 prostate cancer Akt1 , Akt2 , Akt3 , Ccnd1 , Cdk4 , Cdkn2a , E2f1 , E2f3 , Egfr , Erbb2 , Fhit , Foxo3 , Grb2 , Kras , Nras , Pdpk1 , Pik3ca , Pik3cb , Pik3r1 , Pik3r2 , Pten , Raf1 , Rassf1 , Rb1 , Sos1 , Tp53 Prostate Cancer, Hereditary, 1 Braf , Hras , Kras , Nras , Pten , Tp53 Prostate Cancer, Somatic Pten prostate carcinoma Akt2 , Bad , Met , Prkca prostate carcinoma in situ Ccnd1 , Kras , Pten , Rxra prostatic hypertrophy Bad , Cdk4 , Pten Prostatic Neoplasms Akt1 , Akt2 , Bad , Braf , Casp9 , Ccnd1 , Cdkn2a , Egf , Egfr , Erbb2 , Fhit , Hras , Kras , Map2k1 , Map2k2 , Mapk3 , Met , Pik3ca , Pik3cb , Pik3cd , Pik3r1 , Pik3r2 , Pten , Raf1 , Rassf1 , Rb1 , Rxra , Tgfa , Tp53 proteasome-associated autoinflammatory syndrome 1 Rxrb Proteus syndrome Akt1 , Pten Pseudomonas Infections Pten psoriasis Tgfa , Tp53 PTEN hamartoma tumor syndrome Pik3ca , Pten pterygium Tp53 ptosis Sos1 Pulmonary Arterial Hypertension Fhit pulmonary edema Tgfa pulmonary fibrosis Bad , Egf , Egfr , Tgfa pulmonary hypertension Egfr , Mapk1 , Mapk3 , Tgfa , Tp53 Pulmonary Hypertension, Hypoxia-Induced Egfr pulmonary tuberculosis Akt1 , Egfr pulmonary valve stenosis Braf , Hras , Sos1 Pyogenic Granuloma Nras Rafiq syndrome Rxra RASopathy Braf , Hras , Kras , Map2k1 , Map2k2 , Nras , Raf1 , Sos1 , Sos2 rectal benign neoplasm Egfr , Nras , Pik3ca , Pik3cb Recurrence Erbb2 , Hras , Kras Recurrent Infections, with Encephalopathy, Hepatic Dysfunction, and Cardiovascular Malformations Ccnd1 renal carcinoma Met renal cell carcinoma Akt1 , Akt3 , Bad , Ccnd1 , Cdk4 , Cdkn2a , Egfr , Erbb2 , Fhit , Kras , Map2k1 , Mapk1 , Met , Nras , Pik3ca , Pik3r1 , Pten , Rassf1 , Rb1 , Tp53 Renal Cell Carcinoma 1 Met , Nras , Pik3ca , Pten , Tp53 renal fibrosis Kras , Mapk1 renal hypomagnesemia 3 Egf renal hypomagnesemia 4 Egf renal hypomagnesemia 5 with ocular involvement Egf Reperfusion Injury Akt1 , Bad , Casp9 , Ccnd1 , Egf , Met , Pik3r1 , Prkca , Pten , Tp53 Retina Reperfusion Injury Bad retinal detachment Casp9 retinitis pigmentosa Prkcg , Pten retinoblastoma Cdkn2a , Rb1 rhabdomyosarcoma Cdkn2a , Hras , Pten , Tp53 rheumatoid arthritis Cdk6 , Foxo3 , Pik3r1 Right Ventricular Hypertrophy Akt1 , Egfr , Prkca , Tgfa Roifman-Chitayat Syndrome Pik3cd rosette-forming glioneuronal tumor Pik3ca Salivary Gland Neoplasms Ccnd1 , Hras SAPHO syndrome Tp53 sarcoma Met , Pik3ca , Tp53 sarcomatoid mesothelioma Cdkn2a schistosomiasis Rassf1 schizophrenia Akt1 , Cdkn2a , Egf , Egfr , Fhit , Mapk3 , Met , Pik3cb , Pik3cd , Pten , Tp53 sciatic neuropathy Rarb , Rxra scrapie Casp9 Sebaceous Nevus Syndrome and Hemimegalencephaly Hras , Kras , Nras seborrheic keratosis Pik3ca seminoma Fhit , Hras , Kras , Rb1 Senior-Loken syndrome Akt3 Senior-Loken Syndrome 7 Akt3 sensorineural hearing loss Prkcb Sepsis Bad , Casp9 , Mapk1 , Mapk3 , Rxra Septic Peritonitis Pik3r1 , Pten serous cystadenocarcinoma Tp53 severe acute respiratory syndrome Casp9 , Egfr , Mapk1 , Mapk3 severe combined immunodeficiency Stk4 Sezary's disease Braf , Cdkn2a , Mapk1 , Plcg1 , Pten , Rb1 , Tp53 shigellosis Egf Shock Mapk1 , Mapk3 SHORT syndrome Pik3r1 short-rib thoracic dysplasia 9 with or without polydactyly Pdpk1 silicosis Prkcb , Pten sinusitis Met Sjogren's syndrome E2f1 skin benign neoplasm Hras skin disease Pten skin melanoma Akt1 , Braf , Cdk4 , Cdkn2a , Erbb2 , Hras , Kras , Map2k1 , Map2k2 , Nras , Pik3ca , Pik3r1 , Pten , Raf1 , Tp53 Skin Neoplasms Akt1 , Hras , Pten , Tp53 skin squamous cell carcinoma Pten small cell carcinoma Rassf1 , Rb1 , Tp53 small intestine adenocarcinoma Map2k1 , Map2k2 Small-For-Size Syndrome Bad Smith-Kingsmore Syndrome Pik3r1 , Pik3r2 Smith-Magenis syndrome Map2k2 spastic ataxia Pik3r5 spermatocytoma Hras Spinal Cord Injuries Akt1 , Casp9 , Egfr , Foxo3 , Met , Prkcg , Pten , Tp53 Spinal Cord Reperfusion Injury Bad spinocerebellar ataxia type 14 Prkcg Spinocerebellar Ataxias Prkcg Splenic Neoplasms Hras Splenomegaly Kras , Nras spondyloarthropathy Tp53 spondylocostal dysostosis 5 Mapk3 sporadic breast cancer Pten Sporadic Papillary Renal Cell Carcinoma Braf , Cdkn2a , Erbb2 , Hras , Pik3ca , Tp53 squamous cell carcinoma Akt1 , Ccnd1 , Cdkn2a , Egfr , Fhit , Hras , Kras , Mapk1 , Mapk3 , Met , Pik3ca , Pten , Rarb , Rb1 , Tp53 Staphylococcal Pneumonia Akt1 status epilepticus Ccnd1 , Foxo3 , Prkcb steatotic liver disease Akt1 , Met , Pten Stevens-Johnson syndrome Rb1 stomach cancer Casp9 , Cdk4 , Cdkn2a , Egfr , Erbb2 , Kras , Pik3ca , Pten , Tp53 stomach carcinoma Erbb2 , Kras stomach disease Pten Stomach Neoplasms Ccnd1 , Cdk4 , Cdkn2a , Egfr , Erbb2 , Fhit , Hras , Kras , Mapk1 , Mapk3 , Met , Nras , Pik3ca , Prkcb , Rarb , Rxrb , Tgfa , Tp53 Streptococcus pneumonia Pik3cd Stroke Akt1 , Cdk6 , Pik3ca , Raf1 , Sos1 , Tgfa Sturge-Weber syndrome Map2k1 Subarachnoid Hemorrhage Casp9 , Pik3cb substance-induced psychosis Akt1 substance-related disorder Fhit , Pten Sudden Unexpected Nocturnal Death Syndrome Sos1 syndromic microphthalmia 12 Rarb systemic lupus erythematosus Rassf5 , Rxra T-cell acute lymphoblastic leukemia Cdkn2a , Kras T-Cell Immunodeficiency, Recurrent Infections, and Autoimmunity with or without Cardiac Malformations Stk4 T-cell non-Hodgkin lymphoma Akt1 , Hras , Tp53 temporal lobe epilepsy Foxo3 teratoma Kras Testis Reperfusion Injury Casp9 , Mapk1 , Mapk3 tethered spinal cord syndrome Braf thrombosis Pik3cb thymoma Cdkn2a , Hras , Tp53 Thymus Neoplasms Tp53 thyroid cancer Mapk1 Thyroid Carcinoma, Nonmedullary 1 Kras thyroid gland anaplastic carcinoma Tp53 thyroid gland follicular carcinoma Kras , Pten thyroid gland medullary carcinoma Kras thyroid gland papillary carcinoma Braf , Ccnd1 , Rxrg , Tp53 Thyroid Neoplasms Akt1 , Akt2 , Braf , Ccnd1 , Hras , Kras , Mapk1 , Nras , Pik3ca , Pten , Rxra , Tp53 Tobacco Use Disorder Egfr Tongue Neoplasms Cdkn2a , Hras , Tp53 tongue squamous cell carcinoma Ccnd1 , Egfr , Met , Tp53 toxic shock syndrome Pik3cd transient cerebral ischemia Bad , Casp9 , Cdkn2a , Pten transitional cell carcinoma Akt1 , Braf , Ccnd1 , Cdk4 , Cdkn2a , Egfr , Erbb2 , Hras , Kras , Map2k1 , Mapk1 , Nras , Pik3ca , Pten , Rassf1 , Rxra , Tp53 transthyretin amyloidosis Tp53 traumatic brain injury Bad , Tp53 Trichilemmoma Hras Trigeminal Nerve Injuries Tp53 trigeminal neuralgia Mapk1 , Mapk3 , Prkcg trilateral retinoblastoma Rb1 Triple Negative Breast Neoplasms Cdk4 , Cdk6 , Kras Trisomy Cdkn2a tuberous sclerosis 1 Rxra Tympanic Membrane Perforation Egf type 1 diabetes mellitus Casp9 , Cdk4 , E2f1 , Egfr type 2 diabetes mellitus Akt1 , Akt2 , Bad , Braf , Casp9 , Cdk4 , Egfr , Mapk1 , Mapk3 , Pik3cb , Pik3r1 , Prkcb ulcerative colitis Tp53 ureteral obstruction Akt1 , Casp9 , Mapk1 , Mapk3 urethral obstruction Mapk1 urinary bladder cancer Akt1 , Casp9 , Ccnd1 , Cdk4 , Cdkn2a , E2f1 , Egfr , Erbb2 , Fhit , Hras , Kras , Pik3ca , Prkca , Pten , Rassf1 , Rb1 , Tp53 Urinary Bladder Neoplasm Cdkn2a , Hras , Kras , Rb1 , Tp53 Urinary Incontinence Mapk1 Urogenital Neoplasms Cdkn2a , Tp53 Urologic Neoplasms Ccnd1 , Erbb2 , Fhit , Tp53 uterine cancer Braf , Erbb2 , Hras , Kras , Map2k1 , Nras , Pik3ca , Pik3r1 , Pten , Sos1 , Tp53 uterine carcinosarcoma Cdkn2a , Erbb2 , Hras , Pik3ca , Pik3r1 , Pten , Tp53 Uterine Cervical Neoplasms Akt1 , Ccnd1 , Cdk4 , Cdkn2a , Egfr , Erbb2 , Fhit , Hras , Kras , Mapk1 , Pik3ca , Pten , Rarb , Tp53 uterine corpus cancer Pten Uterine Neoplasms Kras varicocele Casp9 vascular dementia Mapk1 , Tp53 Vascular Malformations Braf , Kras , Map2k1 , Pik3ca , Pik3r1 Vascular Remodeling Egfr velocardiofacial syndrome Mapk1 Ventilator-Induced Lung Injury Akt1 , Erbb2 Ventricular Dysfunction, Left Pik3ca Ventricular Fibrillation Egfr ventricular septal defect Braf Ventricular Tachycardia Raf1 , Sos1 very long chain acyl-CoA dehydrogenase deficiency Tp53 viral encephalitis E2f1 visceral leishmaniasis Pik3cd visual epilepsy Bad , Ccnd1 , Pten vitiligo Pik3r1 von Hippel-Lindau disease Ccnd1 vulva cancer Akt1 Vulvar Lichen Sclerosus Cdkn2a , Tp53 Vulvar Neoplasms Cdkn2a , Fhit , Tp53 Wallerian Degeneration Erbb2 , Rxra Weight Gain Pik3ca Weissenbacher-Zweymuller syndrome Mapk1 withdrawal disorder Mapk1 , Mapk3 Wounds and Injuries Egf X-linked agammaglobulinemia Pik3r1 X-linked VACTERL association Pten xeroderma pigmentosum Tp53 Zollinger-Ellison syndrome Egf